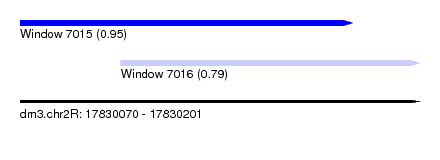
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,830,070 – 17,830,201 |
| Length | 131 |
| Max. P | 0.947618 |

| Location | 17,830,070 – 17,830,179 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.81 |
| Shannon entropy | 0.39487 |
| G+C content | 0.54271 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -18.00 |
| Energy contribution | -19.25 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
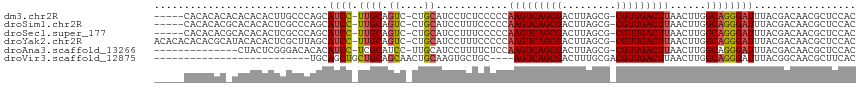
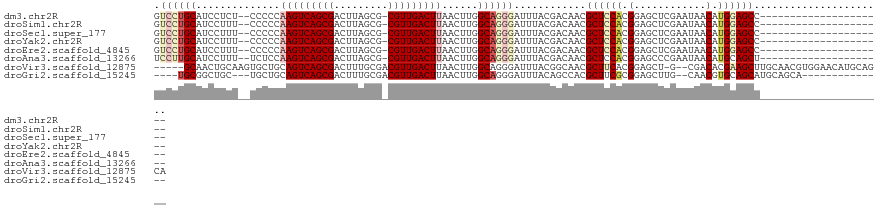
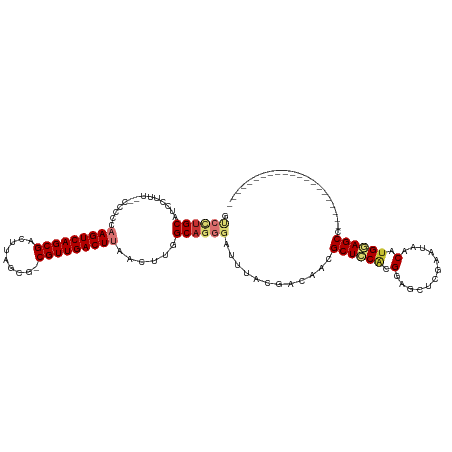
>dm3.chr2R 17830070 109 + 21146708 -----CACACACACACACACUUGCCCAGCAUCC-UUGCAGUC-CUGCAUCCUCUCCCCCAAGUCAGCGACUUAGCG-CGUUGACUUAACUUGGCAGGGAUUUACGACAACGCUCCAC -----.................((...(((...-.))).(((-.((.(((((..((...(((((((((........-))))))))).....))..))))).)).)))...))..... ( -26.30, z-score = -1.95, R) >droSim1.chr2R 16465788 109 + 19596830 -----CACACACGCACACACUCGCCCAGCAUCC-UUGCAGUC-CUGCAUCCUUUCCCCCAAGUCAGCGACUUAGCG-CGUUGACUUAACUUGGCAGGGAUUUACGACAACGCUCCAC -----.......((.............(((...-.))).(((-.((.((((((.((...(((((((((........-))))))))).....)).)))))).)).)))...))..... ( -28.50, z-score = -2.41, R) >droSec1.super_177 14036 109 + 33666 -----CACACACGCACACACUCGCCCAGCAUCC-UUGCAGUC-CUGCAUCCUUUCCCCCAAGUCAGCGACUUAGCG-CGUUGACUUAACUUGGCAGGGAUUUACGACAACGCUCCAC -----.......((.............(((...-.))).(((-.((.((((((.((...(((((((((........-))))))))).....)).)))))).)).)))...))..... ( -28.50, z-score = -2.41, R) >droYak2.chr2R 9785831 114 - 21139217 ACACACACACGCAUACACACUCGCUUAGCAUCC-UUGCAGUC-CUGCAUCCUUUCCCCCAAGUCAGCGACUUAGCG-CGUUGACUUAACUUGGCAGGGAUUUACGACAACGCUCCAC ..........((...............(((...-.))).(((-.((.((((((.((...(((((((((........-))))))))).....)).)))))).)).)))...))..... ( -28.50, z-score = -2.56, R) >droAna3.scaffold_13266 8681730 100 - 19884421 --------------CUACUCGGGACACACAUCC-UCGCAUCC-UUGCAUCCUUUUCUCCAAGUCAGCGACUUAGCG-CGUUGACUUAACUUGGCAGGGAUUUACGACAACGCUCCAC --------------......((..(........-..(((...-.)))((((((.((...(((((((((........-))))))))).....)).))))))..........)..)).. ( -24.00, z-score = -1.63, R) >droVir3.scaffold_12875 10418995 87 + 20611582 --------------------------UGCAGCUGCUGCAGCAACUGCAAGUGCUGC----AGUCAGCGACUUUGCGACGUUGACUUAACUUGGCAGGGAUUUACGGCAACGCUUCAC --------------------------(((.((((((((((((........))))))----)).))))..((((((.(.((((...)))).).)))))).......)))......... ( -32.10, z-score = -1.34, R) >consensus _____CACACAC_CACACACUCGCCCAGCAUCC_UUGCAGUC_CUGCAUCCUUUCCCCCAAGUCAGCGACUUAGCG_CGUUGACUUAACUUGGCAGGGAUUUACGACAACGCUCCAC ...........................((.......((((...))))((((((.((...(((((((((.(.....).))))))))).....)).))))))..........))..... (-18.00 = -19.25 + 1.25)
| Location | 17,830,103 – 17,830,201 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 80.03 |
| Shannon entropy | 0.35456 |
| G+C content | 0.54301 |
| Mean single sequence MFE | -32.99 |
| Consensus MFE | -20.39 |
| Energy contribution | -21.49 |
| Covariance contribution | 1.10 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.793215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
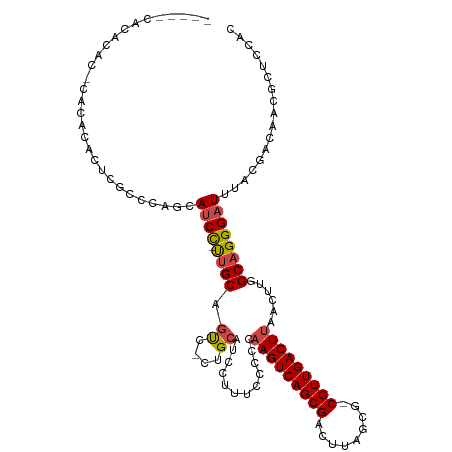
>dm3.chr2R 17830103 98 + 21146708 GUCCUGCAUCCUCU--CCCCCAAGUCAGCGACUUAGCG-CGUUGACUUAACUUGGCAGGGAUUUACGACAACGCUCCACGGAGCUCGAAUAACAUGGAGCC--------------------- (((.((.(((((..--((...(((((((((........-))))))))).....))..))))).)).)))...((((((.(............).)))))).--------------------- ( -31.10, z-score = -2.09, R) >droSim1.chr2R 16465821 98 + 19596830 GUCCUGCAUCCUUU--CCCCCAAGUCAGCGACUUAGCG-CGUUGACUUAACUUGGCAGGGAUUUACGACAACGCUCCACGGAGCUCGAAUAACAUGGAGCC--------------------- (((.((.((((((.--((...(((((((((........-))))))))).....)).)))))).)).)))...((((((.(............).)))))).--------------------- ( -32.10, z-score = -2.57, R) >droSec1.super_177 14069 98 + 33666 GUCCUGCAUCCUUU--CCCCCAAGUCAGCGACUUAGCG-CGUUGACUUAACUUGGCAGGGAUUUACGACAACGCUCCACGGAGCUCGAAUAACAUGGAGCC--------------------- (((.((.((((((.--((...(((((((((........-))))))))).....)).)))))).)).)))...((((((.(............).)))))).--------------------- ( -32.10, z-score = -2.57, R) >droYak2.chr2R 9785869 98 - 21139217 GUCCUGCAUCCUUU--CCCCCAAGUCAGCGACUUAGCG-CGUUGACUUAACUUGGCAGGGAUUUACGACAACGCUCCACGGAGCUCGAAUAACAUGGAGCC--------------------- (((.((.((((((.--((...(((((((((........-))))))))).....)).)))))).)).)))...((((((.(............).)))))).--------------------- ( -32.10, z-score = -2.57, R) >droEre2.scaffold_4845 11962482 98 + 22589142 GUCCUGCAUCCUUU--CCCCCAAGUCAGCGACUUAGCG-CGUUGACUUAACUUGGCAGGGAUUUACGACAACGCUCCACGGAGCUCGAAUAACAUGGAGCC--------------------- (((.((.((((((.--((...(((((((((........-))))))))).....)).)))))).)).)))...((((((.(............).)))))).--------------------- ( -32.10, z-score = -2.57, R) >droAna3.scaffold_13266 8681754 98 - 19884421 UCCUUGCAUCCUUU--UCUCCAAGUCAGCGACUUAGCG-CGUUGACUUAACUUGGCAGGGAUUUACGACAACGCUCCACGGAGCCCGAAUAACAUGCAGCU--------------------- ...((((((((((.--((...(((((((((........-))))))))).....)).)))))...........((((....))))..........)))))..--------------------- ( -26.60, z-score = -1.37, R) >droVir3.scaffold_12875 10419008 114 + 20611582 -----GCAACUGCAAGUGCUGCAGUCAGCGACUUUGCGACGUUGACUUAACUUGGCAGGGAUUUACGGCAACGCUUCACGGAGCU-G--CGACACGAAGCUUGCAACGUGGAACAUGCAGCA -----((....))...(((((((.(((((..((((((.(.((((...)))).).))))))......(....)))).((((..((.-(--(........))..))..))))))...))))))) ( -36.60, z-score = -0.07, R) >droGri2.scaffold_15245 3567039 99 + 18325388 ----UGCGGCUGC---UGCUGCAGUCAGCGACUUUGCGACGUUGACUUAACUUGGCAGGGAUUUACAGCCACGCUUCGCGGAGCUUG--CAACGUGCAGCAUGCAGCA-------------- ----....(((((---(((((((((..((((((((((((.((((...)))).((((.(.......).))))....)))))))).)))--).)).))))))).))))).-------------- ( -41.20, z-score = -1.23, R) >consensus GUCCUGCAUCCUUU__CCCCCAAGUCAGCGACUUAGCG_CGUUGACUUAACUUGGCAGGGAUUUACGACAACGCUCCACGGAGCUCGAAUAACAUGGAGCC_____________________ .((((((..............(((((((((.(.....).)))))))))......))))))............((((((.(............).))))))...................... (-20.39 = -21.49 + 1.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:42:04 2011