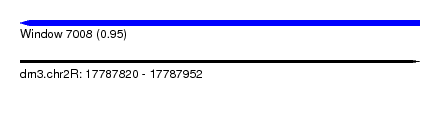
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,787,820 – 17,787,952 |
| Length | 132 |
| Max. P | 0.953846 |

| Location | 17,787,820 – 17,787,952 |
|---|---|
| Length | 132 |
| Sequences | 11 |
| Columns | 152 |
| Reading direction | reverse |
| Mean pairwise identity | 65.73 |
| Shannon entropy | 0.68417 |
| G+C content | 0.47353 |
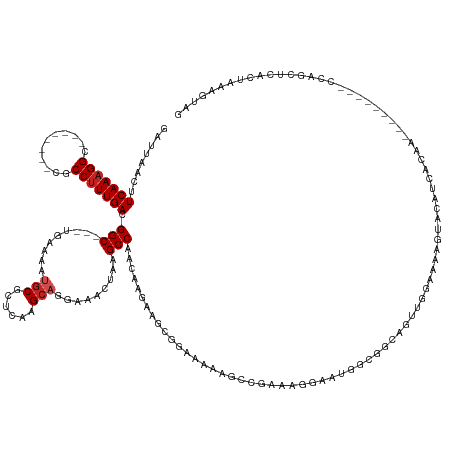
| Mean single sequence MFE | -37.07 |
| Consensus MFE | -13.82 |
| Energy contribution | -14.18 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.953846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
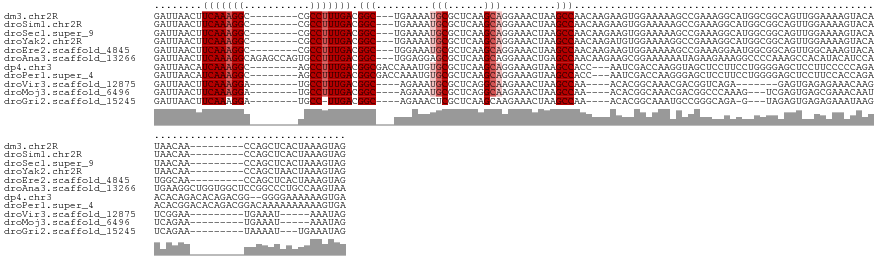
>dm3.chr2R 17787820 132 - 21146708 GAUUAACUUCAAAGGC--------CGCCUUUGACGGC---UGAAAAUGCGCUCAAGCAGGAAACUAAGCCAACAAGAAGUGGAAAAAGCCGAAAGGCAUGGCGGCAGUUGGAAAAGUACAUAACAA---------CCAGCUCACUAAAGUAG .....((((.....((--------((((((((..(((---(......((......))((....)).))))..))))...........(((....)))..))))))((((((...............---------)))))).....)))).. ( -40.36, z-score = -2.95, R) >droSim1.chr2R 16423328 132 - 19596830 GAUUAACUUCAAAGGC--------CGCCUUUGACGGC---UGAAAAUGCGCUCAAGCAGGAAACUAAGCCAACAAGAAGUGGAAAAAGCCGAAAGGCAUGGCGGCAGUUGGAAAAGUACAUAACAA---------CCAGCUCACUAAAGUAG .....((((.....((--------((((((((..(((---(......((......))((....)).))))..))))...........(((....)))..))))))((((((...............---------)))))).....)))).. ( -40.36, z-score = -2.95, R) >droSec1.super_9 1102959 132 - 3197100 GAUUAACUUCAAAGGC--------CGCCUUUGACGGC---UGAAAAUGCGCUCAAGCAGGAAACUAAGCCAACAAGAAGUGGAAAAAGCCGAAAGGCAUGGCGGCAGUUGGAAAAGUACAUAACAA---------CCAGCUCACUAAAGUAG .....((((.....((--------((((((((..(((---(......((......))((....)).))))..))))...........(((....)))..))))))((((((...............---------)))))).....)))).. ( -40.36, z-score = -2.95, R) >droYak2.chr2R 9742608 132 + 21139217 GAUUAACUUCAAAGGC--------CGCCUUUGACGGC---UGAAAAUGCGCUCAAGCAGGAAACUAAGCCAACAAGAUGUGGAAAAGGCCGAAAGGCAUGGCGGCAGUUGGAAAAGUACAUAACAA---------CCAGCUAACUAAAGUAG .....((((.....((--------((((((((..(((---(......((......))((....)).))))..))))...........(((....)))..))))))((((((...............---------)))))).....)))).. ( -40.76, z-score = -2.93, R) >droEre2.scaffold_4845 11921534 132 - 22589142 GAUUAACUUCAAAGGC--------CGCCUUUGACGGC---UGGAAAUGCGCUCAAGCAGGAAACUAAGCCAACAAGAAGUGGAAAAAGCCGAAAGGAAUGGCGGCAGUUGGCAAAGUACAUGGCAA---------CCAGCUCACUAAAGUAG .....((((((((((.--------..))))))).(((---(((...(((((....))((....))..((((((......(....)..((((..(....)..)))).))))))..........))).---------))))))......))).. ( -37.60, z-score = -1.04, R) >droAna3.scaffold_13266 11166077 149 - 19884421 GAUUAACUUCAAAGGCAGAGCCAGUGCCUUUGACGGC---UGGAGGAGCGCUCAAGCAGGAAACUGAGCCAACAAGAAGCGGAAAAAAUAGAAGAAAGGCCCCAAAGCCACAUACAUCCAUGAAGGCUGGUGGCUCCGGCCCUGCCAAGUAA .............(((((.(((...(((......)))---....((((((((....(((....)))((((...........................(((......))).(((......)))..))))))).)))))))).)))))...... ( -46.90, z-score = -0.84, R) >dp4.chr3 15343189 139 - 19779522 GAUUAACAUCAAAGGC--------AGCCUUUGACGGCGACCAAAUGUGCGCUCAAGCAGGAAAGUAAGCCACC---AAUCGACCAAGGUAGCUCCUUCCUGGGGAGCUCCUUCCCCCAGAACACAGACACAGACGG--GGGGAAAAAAGUGA ((((....(((((((.--------..))))))).(((.......(.(((......))).).......)))...---))))......((.(((((((.....)))))))))(((((((..................)--))))))........ ( -42.11, z-score = -0.68, R) >droPer1.super_4 3454075 141 + 7162766 GAUUAACAUCAAAGGC--------AGCCUUUGACGGCGACCAAAUGUGCGCUCAAGCAGGAAAGUAAGCCACC---AAUCGACCAAGGGAGCUCCUUCCUGGGGAGCUCCUUCCACCAGAACACGGACACAGACGGACAAAAAAAAAAGUGA ((((....(((((((.--------..))))))).(((.......(.(((......))).).......)))...---))))..((.(((((((((((.....)))))))))))...((.......))........))................ ( -38.74, z-score = -1.03, R) >droVir3.scaffold_12875 10366853 115 - 20611582 GAUUAACUUCAAAGGA--------UGCCUUUGACGGC----AGAAAUGCGCUCAGGCAAGAAACUAAGCCAA----ACACGGCAAACGACGGUCAGA-------GAGUGAGAGAAACAAGUCGGAA---------UGAAAU-----AAAUAG ((((..(.(((((((.--------..))))))).)((----(....)))((((.(((..........)))..----....(((........)))...-------))))..........))))....---------......-----...... ( -23.20, z-score = -0.75, R) >droMoj3.scaffold_6496 10261116 119 + 26866924 GAUUAACUUCAAAGGA--------UGCCUUUGACGGC----AGAAAUGCGCUCAGGCAAGAAACUAAGCCAA----ACACGGCAAACGACGGCCCAAAG---UCGAGUGAGCGAAACAAUUCAGAA---------UGAAAU-----AAAUAG ......(.(((((((.--------..))))))).)((----(....)))(((((.............(((..----....)))...((((........)---)))..))))).......(((....---------.)))..-----...... ( -29.30, z-score = -2.24, R) >droGri2.scaffold_15245 3514239 119 - 18325388 GAUUAACUUCAAAGGA--------UGCC-UUGACGGC----AGAAACUCGCUCAAGCAAGAAACUAAGCCAA----ACACGGCAAAUGCCGGGCAGA-G---UAGAGUGAGAGAAAUAAGUCAGAA---------UAAAAU---UGAAAUAG ((((.(.(((......--------((((-.....)))----)....(((((((.........(((..(((..----...((((....)))))))..)-)---).))))))).))).).))))....---------......---........ ( -28.10, z-score = -2.29, R) >consensus GAUUAACUUCAAAGGC________CGCCUUUGACGGC___UGAAAAUGCGCUCAAGCAGGAAACUAAGCCAACAAGAAGCGGAAAAAGCCGAAAGGAAUGGCGGCAGUUGGAAAAGUACAUCACAA_________CCAGCUCACUAAAGUAG ........(((((((...........))))))).(((.........(((......))).........))).................................................................................. (-13.82 = -14.18 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:41:58 2011