| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,786,895 – 17,786,989 |
| Length | 94 |
| Max. P | 0.961361 |

| Location | 17,786,895 – 17,786,989 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 68.30 |
| Shannon entropy | 0.63682 |
| G+C content | 0.35019 |
| Mean single sequence MFE | -19.56 |
| Consensus MFE | -12.43 |
| Energy contribution | -12.35 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.26 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.961361 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
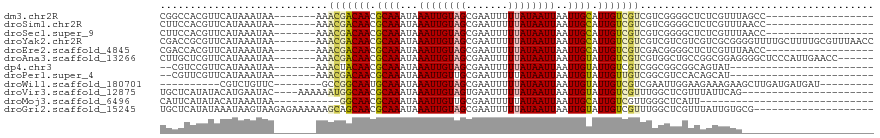
>dm3.chr2R 17786895 94 + 21146708 CGGCCACGUUCAUAAAUAA-------AAACGACAACGCAAAUAAAUUGUAGCGAAUUUUUAUAAUUAAUUGCAUUGUCGUCGUCGGGGCUCUCGUUUAGCC------------------ ((((.(((((.........-------.)))(((((.((((...((((((((.......))))))))..)))).))))))).)))).((((.......))))------------------ ( -20.60, z-score = -1.05, R) >droSim1.chr2R 16422407 94 + 19596830 CUUCCACGUUCAUAAAUAA-------AAACGACAACGCAAAUAAAUUGUAGCGAAUUUUUAUAAUUAAUUGCAUUGUCGUCGUCGGGGCUCUCGUUUAACC------------------ .(((((((...........-------..(((((((.((((...((((((((.......))))))))..)))).)))))))))).)))).............------------------ ( -17.91, z-score = -1.31, R) >droSec1.super_9 1102029 94 + 3197100 CUUCCACGUUCAUAAAUAA-------AAACGACAACGCAAAUAAAUUGUAGCGAAUUUUUAUAAUUAAUUGCAUUGUCGUCGUCGGGGCUCUCGUUUAACC------------------ .(((((((...........-------..(((((((.((((...((((((((.......))))))))..)))).)))))))))).)))).............------------------ ( -17.91, z-score = -1.31, R) >droYak2.chr2R 9741622 112 - 21139217 CGACCGCGUUCAUAAAUAA-------AAACGACAACGCAAAUAAAUUGUAGCGAAUUUUUAUAAUUAAUUGCAUUGUCGUCGUCGUCGUCGUCGCGGGGUUUUGCUUUUGCGUUUAACC ...(((((...........-------..(((((((.((((...((((((((.......))))))))..)))).)))))))............)))))((((.(((....)))...)))) ( -29.51, z-score = -1.14, R) >droEre2.scaffold_4845 11920659 94 + 22589142 CGACCACGUUCAUAAAUAA-------AAACGACAACGCAAAUAAAUUGUAGCGAAUUUUUAUAAUUAAUUGCAUUGUCGUCGACGGGGCUCUCGUUUAACC------------------ .((((.((((.........-------..(((((((.((((...((((((((.......))))))))..)))).))))))).)))).)).))..........------------------ ( -19.30, z-score = -1.17, R) >droAna3.scaffold_13266 11165328 106 + 19884421 CUUGCUCGUUCAUAAAUAA-------AAACGACAACGCAAAUAAAUUGUAGCGAAUUUUUAUAAUUAAUUGUAUUGUCGUCGUGGCUGCCGGCGGAGGGGCUCCCAUUGAACC------ .......(((((.......-------..(((((((..(((...((((((((.......))))))))..)))..))))))).((((..(((........)))..))))))))).------ ( -27.70, z-score = -1.51, R) >dp4.chr3 15342394 86 + 19779522 --CGUCCGUUCAUAAAUAA-------AAACUACAACGCAAAUAAAUUGUAGCGAAUUUUUAUAAUUAAUUGUAUUGUCGUCGGCGGCGGCAGUAU------------------------ --.............((((-------(((((((((..........))))))....)))))))........((((((((((.....))))))))))------------------------ ( -16.10, z-score = -0.39, R) >droPer1.super_4 3453317 86 - 7162766 --CGUUCGUUCAUAAAUAA-------AAACGACAACGCAAAUAAAUUGUUGCGAAUUUUUAUAAUUAAUUGUAUUGUUGUCGGCGUCCACAGCAU------------------------ --.................-------...(((((((((((........)))).......(((((....)))))..)))))))((.......))..------------------------ ( -15.20, z-score = -0.25, R) >droWil1.scaffold_180701 3111781 92 + 3904529 ----------CGUCUGUUC--------GCCGGCAAUGCAAAUAAAUUGUAGCGAAUUUUUAUAAUUAAUUGUAUUGUCGUCGAAUUGGAAGAAAGAAGCUUGAUGAUGAU--------- ----------((((.((((--------(.(((((((((((...((((((((.......))))))))..))))))))))).)))))...(((.......))))))).....--------- ( -23.20, z-score = -2.12, R) >droVir3.scaffold_12875 10365294 93 + 20611582 UGCUCAUAUACAUGAAUAC----AAAAAAUGGCAACGCAAAUAAAUUGUAGUGAAUUUUUAUAAUUAAUUGUAUUGUCGUUUGGCUCGUUUAUUCAG---------------------- ............((((((.----...(((((((((..(((...((((((((.......))))))))..)))..)))))))))........)))))).---------------------- ( -18.00, z-score = -1.37, R) >droMoj3.scaffold_6496 10259216 79 - 26866924 CAUUCAUAUACAUAAAUAA-----------GGCAACGCAAAUAAAUUGUUGCGAAUUUUUAUAAUUAAUUGCAUUGUCGUUGGGCUCAUU----------------------------- ...................-----------(((((.((((...((((((...........))))))..)))).)))))............----------------------------- ( -10.00, z-score = 0.75, R) >droGri2.scaffold_15245 3512465 99 + 18325388 UGCUCAUAUAAAUAAGUAAGAGAAAAAAGCAGCAACGCAAAUAAAUUGUAGCGAAUUUUUAUAAUUAAUUGUAUUGUCGUUUGGCUCGUUUAUUGUGCG-------------------- .((.((.(((((..(((.........((((.((((..(((...((((((((.......))))))))..)))..)))).)))).)))..))))))).)).-------------------- ( -19.30, z-score = -0.56, R) >consensus CGCCCACGUUCAUAAAUAA_______AAACGACAACGCAAAUAAAUUGUAGCGAAUUUUUAUAAUUAAUUGCAUUGUCGUCGGCGUCGCUCUCGUUG______________________ ............................(((((((.((((...((((((((.......))))))))..)))).)))))))....................................... (-12.43 = -12.35 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:41:57 2011