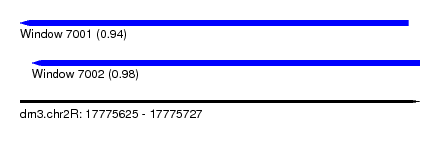
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,775,625 – 17,775,727 |
| Length | 102 |
| Max. P | 0.979357 |

| Location | 17,775,625 – 17,775,724 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 65.64 |
| Shannon entropy | 0.71406 |
| G+C content | 0.55685 |
| Mean single sequence MFE | -29.36 |
| Consensus MFE | -15.50 |
| Energy contribution | -16.29 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.68 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.940981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
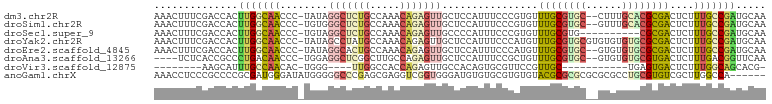
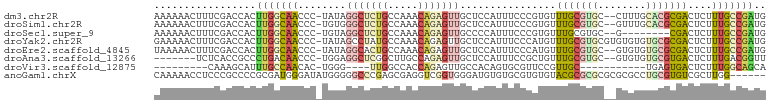
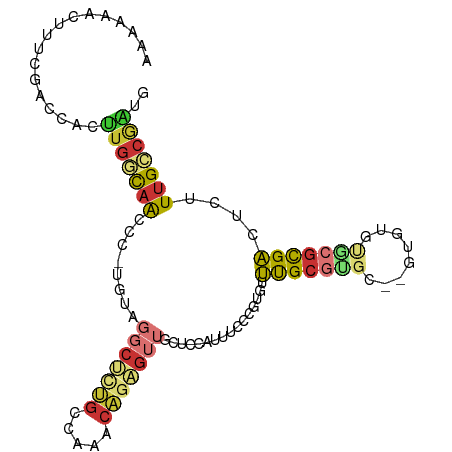
>dm3.chr2R 17775625 99 - 21146708 AAACUUUCGACCACUUGGCAACCC-UAUAGGCUCUGCCAAACAGAGUUGCUCCAUUUCCCGUGUUUGCGUGC--CUUUGCACGCGACUCUUUGCCGAUGCAA ..............(((((((...-....(((((((.....)))))))................((((((((--....))))))))....)))))))..... ( -27.50, z-score = -2.13, R) >droSim1.chr2R 16411049 99 - 19596830 AAACUUUCGACCACUUGGCAACCC-UGUGGGCUCUGCCAAACAGAGUUGCUCCAUUUCCCGUGUUUGCGUGC--GUUUGCACGCGACUCUUUGCCGAUGCAA ..............(((((((...-.((((((((((.....))))))....)))).........((((((((--....))))))))....)))))))..... ( -29.30, z-score = -1.11, R) >droSec1.super_9 1090704 91 - 3197100 AAACUUUCGACCACUUGGCAACCC-UGUAGGCUCUGCCAAACAGAGUUGCCCCAUUUCCCGUGUUUGCGUG----------CGCGACUCUUUGCCGAUGCAA ......(((.......((((((.(-(((.(((...)))..)))).))))))........(((((......)----------)))).........)))..... ( -24.10, z-score = -1.14, R) >droYak2.chr2R 9730000 101 + 21139217 AAACUUUCGACCACUUGGCAACCC-UAUAGCCUAUGCCAAACAGAGUUGCUCCAUUUCCCAUGUUUGCGUGCGUGUGUGUGCGCGACUCUUUGCCGAUGCAA ..............(((((((...-....((....)).....((((((((.((((....(((((......))))).))).).)))))))))))))))..... ( -25.10, z-score = -0.83, R) >droEre2.scaffold_4845 11909393 99 - 22589142 AAACUUUCGACCACUUGGCAACCC-UAUAGGCACUGCCAAACAGAGUUGCUCCAUUUCCCAUGUUUGCGUGC--GUGUGUGCGCGACUCUUUGCCGAUGCAA ..............(((((((...-....(((...)))....((((((((.((((..(.((((....)))).--).))).).)))))))))))))))..... ( -27.90, z-score = -1.21, R) >droAna3.scaffold_13266 11155186 95 - 19884421 ----UCUCACCGCCCUGACAACCC-UGGAGGCUCGGCUUGCCAGAGUUGCUCCAUUUCCGCUGUUUGCGUGC--GUGUGUGCGUGACUCUUUGACGGUUCAA ----....((((....((((((.(-(((((((...)))).)))).)))).))............(..((..(--....)..))..)........)))).... ( -24.00, z-score = 1.42, R) >droVir3.scaffold_12875 10346680 77 - 20611582 --------AAGCAUUUGCCAACAC-UGGG----UUGGCCACCAGAGUUGCCACAGUGCGUUCCGUUGC-----------UGAGUGACUCUUUGGCAGCACG- --------..(((((((.((((.(-(((.----.......)))).)))).)).))))).....(((((-----------..((......))..)))))...- ( -26.90, z-score = -0.84, R) >anoGam1.chrX 12360919 96 + 22145176 AAACCUCCCGCCCCGCGAUGGGAUAUGGGGGCCCGAGCGAGGUCGGUGGGAUGUGUGCGUGUGUACGCGCGCGCGCGCCUGCGUGUCGCUUGGCCA------ ......((((.(((.....)))...))))((((.((((((.(.(.(..((.(((((((((((....)))))))))))))..))).)))))))))).------ ( -50.10, z-score = -1.25, R) >consensus AAACUUUCGACCACUUGGCAACCC_UGUAGGCUCUGCCAAACAGAGUUGCUCCAUUUCCCGUGUUUGCGUGC__GUGUGUGCGCGACUCUUUGCCGAUGCAA ..............(((((((........(((((((.....)))))))................((((((((......))))))))....)))))))..... (-15.50 = -16.29 + 0.79)
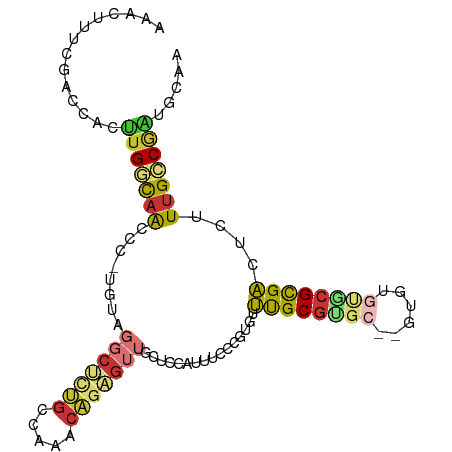
| Location | 17,775,628 – 17,775,727 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 64.50 |
| Shannon entropy | 0.74297 |
| G+C content | 0.54976 |
| Mean single sequence MFE | -29.21 |
| Consensus MFE | -13.65 |
| Energy contribution | -14.62 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.71 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17775628 99 - 21146708 AAAAAACUUUCGACCACUUGGCAACCC-UAUAGGCUCUGCCAAACAGAGUUGCUCCAUUUCCCGUGUUUGCGUGC--CUUUGCACGCGACUCUUUGCCGAUG .................(((((((...-....(((((((.....)))))))................((((((((--....))))))))....))))))).. ( -27.50, z-score = -2.76, R) >droSim1.chr2R 16411052 99 - 19596830 AAAAAACUUUCGACCACUUGGCAACCC-UGUGGGCUCUGCCAAACAGAGUUGCUCCAUUUCCCGUGUUUGCGUGC--GUUUGCACGCGACUCUUUGCCGAUG .................(((((((...-.((((((((((.....))))))....)))).........((((((((--....))))))))....))))))).. ( -29.30, z-score = -1.71, R) >droSec1.super_9 1090707 91 - 3197100 AAAAAACUUUCGACCACUUGGCAACCC-UGUAGGCUCUGCCAAACAGAGUUGCCCCAUUUCCCGUGUUUGCGUGC--G--------CGACUCUUUGCCGAUG .........(((.......((((((.(-(((.(((...)))..)))).))))))........(((((......))--)--------)).........))).. ( -24.10, z-score = -1.79, R) >droYak2.chr2R 9730003 101 + 21139217 GAAAAACUUUCGACCACUUGGCAACCC-UAUAGCCUAUGCCAAACAGAGUUGCUCCAUUUCCCAUGUUUGCGUGCGUGUGUGUGCGCGACUCUUUGCCGAUG .................(((((((...-....((....)).....((((((((.((((....(((((......))))).))).).))))))))))))))).. ( -25.10, z-score = -1.41, R) >droEre2.scaffold_4845 11909396 99 - 22589142 UAAAAACUUUCGACCACUUGGCAACCC-UAUAGGCACUGCCAAACAGAGUUGCUCCAUUUCCCAUGUUUGCGUGC--GUGUGUGCGCGACUCUUUGCCGAUG .................(((((((...-....(((...)))....((((((((.((((..(.((((....)))).--).))).).))))))))))))))).. ( -27.90, z-score = -1.93, R) >droAna3.scaffold_13266 11155189 92 - 19884421 -------UCUCACCGCCCUGACAACCC-UGGAGGCUCGGCUUGCCAGAGUUGCUCCAUUUCCGCUGUUUGCGUGC--GUGUGUGCGUGACUCUUUGACGGUU -------....((((....((((((.(-(((((((...)))).)))).)))).))............(..((..(--....)..))..)........)))). ( -24.00, z-score = 1.26, R) >droVir3.scaffold_12875 10346682 77 - 20611582 ---------CAAAGCAUUUGCCAACAC-UGGG----UUGGCCACCAGAGUUGCCACAGUGCGUUCCGUUGC-----------UGAGUGACUCUUUGGCAGCA ---------....(((((((.((((.(-(((.----.......)))).)))).)).))))).....(((((-----------..((......))..))))). ( -26.90, z-score = -0.71, R) >anoGam1.chrX 12360922 96 + 22145176 CAAAAACCUCCCGCCCCGCGAUGGGAUAUGGGGGCCCGAGCGAGGUCGGUGGGAUGUGUGCGUGUGUACGCGCGCGCGCGCCUGCGUGUCGCUUGG------ ......(((((..(((......)))....))))).((((((((.(.(.(..((.(((((((((((....)))))))))))))..))).))))))))------ ( -48.90, z-score = -1.64, R) >consensus AAAAAACUUUCGACCACUUGGCAACCC_UGUAGGCUCUGCCAAACAGAGUUGCUCCAUUUCCCGUGUUUGCGUGC__GUGUGUGCGCGACUCUUUGCCGAUG .................(((((((........(((((((.....)))))))................(((((((........)))))))....))))))).. (-13.65 = -14.62 + 0.97)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:41:53 2011