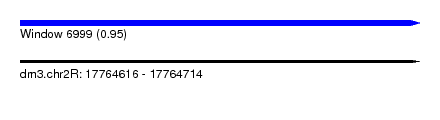
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,764,616 – 17,764,714 |
| Length | 98 |
| Max. P | 0.949229 |

| Location | 17,764,616 – 17,764,714 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.67 |
| Shannon entropy | 0.46032 |
| G+C content | 0.41084 |
| Mean single sequence MFE | -21.96 |
| Consensus MFE | -13.63 |
| Energy contribution | -13.19 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
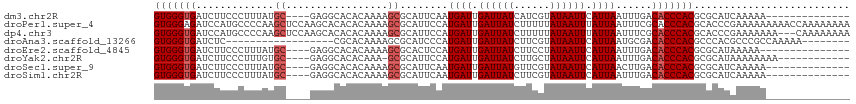
>dm3.chr2R 17764616 98 + 21146708 GUGGGUGAUCUUCCCUUUAUGC----GAGGCACACAAAAGCGCAUUCAAUGAUUGAUUAUCAUCGUAUAAUUCAUUAAUUUGACACCCACGCGCAUCAAAAA-------------- (((((((.........((((((----((.((.(......).))......((((.....)))))))))))).(((......))))))))))............-------------- ( -25.00, z-score = -2.40, R) >droPer1.super_4 3429867 116 - 7162766 GUGGGAGAUCCAUGCCCCAAGCUCCAAGCACACACAAAAGCGCAUUCCAUGAUUGAUUAUCUUUUUAUAAUUUAUUAAUUUCGCACCCACGCACCCGAAAAAAAAACCAAAAAAAA (((((...............((.....))..........(((.......((((.((((((......)))))).))))....))).))))).......................... ( -15.90, z-score = -0.65, R) >dp4.chr3 15318529 113 + 19779522 GUGGGUGAUCCAUGCCCCAAGCUCCAAGCACACACAAAAGCGCAUUCCAUGAUUGAUUAUCUUUUUAUAAUUUAUUAAUUUCGCACCCACGCACCCGAAAAAAA---CAAAAAAAA (((((((....................((.(........).))......((((.((((((......)))))).))))......)))))))..............---......... ( -17.80, z-score = -0.99, R) >droAna3.scaffold_13266 11144599 90 + 19884421 GUGGGUGAUCUC------------------CGCACAAAAGCGCAUCCCAUGAUUGAUUAUCUUCGUAUAAUUCAUUAAUGCGACACCCACGCCCACGCCCGCCAAAAA-------- (((((((.((..------------------.((......))((((....((((.((((((......)))))).)))))))))))))))))..................-------- ( -22.10, z-score = -2.23, R) >droEre2.scaffold_4845 11898148 97 + 22589142 GUGGGUGAUCUUCCCUUUAUGC----GAGGCACACAAAAGCGCACUCCAUGAUUGAUUAUCUUCCUAUAAUUCAUUAAUUUGACACCCACGCGCAUAAAAA--------------- (((((((.((......(((((.----(((((.(......).)).))))))))..((((((......)))))).........)))))))))...........--------------- ( -23.60, z-score = -2.70, R) >droYak2.chr2R 9718471 99 - 21139217 GUGGGUGAUCUUCCCUUUGUGC----GAGGCACACAAA-GCGCAUUCCAUGAUUGAUUAUCUUGCUAUAAUUCAUUAAUUUGACACCCACGCGCAUAAAAAAAA------------ (((((((.((....(((((((.----......))))))-).........((((.((((((......)))))).))))....)))))))))..............------------ ( -23.10, z-score = -1.39, R) >droSec1.super_9 1079753 98 + 3197100 GUGGGUGAUCUUCCCUUUAUGC----GAGGCACACAAAAGCGCAUUCAAUGAUUGAUUAUGUUCGUAUAAUUCAUUAACUUGACACCCACGCGCAUCAAAAA-------------- (((((((.........((((((----((.((........))(((((((.....)))..)))))))))))).(((......))))))))))............-------------- ( -24.10, z-score = -1.81, R) >droSim1.chr2R 16400257 98 + 19596830 GUGGGUGAUCUUCCCUUUAUGC----GAGGCACACAAAAGCGCAUUCAAUGAUUGAUUAUCUUCGUAUAAUUCAUUAAUUUGACACCCACGCGCAUCAAAAA-------------- (((((((.........((((((----(((((.(......).)).......(((.....)))))))))))).(((......))))))))))............-------------- ( -24.10, z-score = -2.19, R) >consensus GUGGGUGAUCUUCCCUUUAUGC____GAGGCACACAAAAGCGCAUUCCAUGAUUGAUUAUCUUCGUAUAAUUCAUUAAUUUGACACCCACGCGCAUCAAAAA______________ (((((((.....((..............))...................((((.((((((......)))))).))))......))))))).......................... (-13.63 = -13.19 + -0.44)
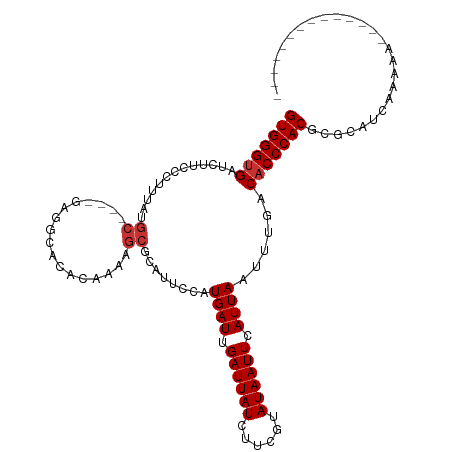
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:41:50 2011