| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,758,096 – 17,758,190 |
| Length | 94 |
| Max. P | 0.989477 |

| Location | 17,758,096 – 17,758,190 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 69.18 |
| Shannon entropy | 0.58903 |
| G+C content | 0.57899 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -22.92 |
| Energy contribution | -22.83 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.989477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17758096 94 + 21146708 AAUUAAUCAUCGUCAUGGGGCGUGUGCACGAUUCGCUUAACGUGCGCCUUUGGCAGUGGUUCCCCCG-----CCCCAUUCCCUUGUUCCACCCAAAAAG---------------- .......((..(..((((((((...(((((..........)))))(((...)))...........))-----))))))..)..))..............---------------- ( -25.40, z-score = -1.23, R) >droSec1.super_9 1073207 92 + 3197100 AAUUAAUCAUCGUCAUGGGGCGUGUGCACGAUUCGCUUAACGUGCGCCUUUGGCGGUGGUUCCCCCG-----CCACACUCCCUUGUUCCAUCUAAAA------------------ ..............((((((((.(((((((..........))))))))..((((((.((...)))))-----))).........)))))))......------------------ ( -29.40, z-score = -2.05, R) >droYak2.chr2R 9711341 115 - 21139217 AAUUAAUCAUCGUCAUGGGGCGUGUGCACGAUUCGCUUAACGUGCGCCUUUGGCGGUGGUUCCCCCGAAACCCCCCUUCCCCCAACCCCCUAUUCCCUUGUUCGCCCCACCAAAA ...............(((((((.(((((((..........)))))))....((.((.((((.......))))))))..........................)))))))...... ( -31.10, z-score = -1.95, R) >droEre2.scaffold_4845 11891576 102 + 22589142 AAUUAAUCAUCGUCAUGGGGCGUGUGCACGAUUCGCUUAACGUGCGCCUUUGGCGGUGGUUCCCCCG-----CCCCUCUCCCUUGCGCCCCCUCCCC--------CCCACCAAAA ................((((((((((((((..........)))))))....(((((.((...)))))-----))..........)))))))......--------.......... ( -34.50, z-score = -3.38, R) >droAna3.scaffold_13266 11139399 88 + 19884421 AAUUAAUCAUCGUCAUGGGGCGUGUGCACGAUUCGCUUAACGUGCGCCUUUGGCGGUGGUGCCACCAGUCCCCCAGUCCCCCCUGCAC--------------------------- ...........((...((((((.(((((((..........))))))))...(((((((....)))).))).....)))))....))..--------------------------- ( -26.90, z-score = 0.04, R) >dp4.chr3 15311280 78 + 19779522 AAUUAAUCAUCGUCAUGGGGCGUGUGCACGAUUCGCUUAACGUGCGCCUUUGGCGGUGGCCUCUCCGGU---CCGGGCAGC---------------------------------- .......((((((((.((((((..((..............))..))))))))))))))((((.......---..))))...---------------------------------- ( -26.74, z-score = -0.10, R) >droPer1.super_4 3422636 78 - 7162766 AAUUAAUCAUCGUCAUGGGGCGUGUGCACGAUUCGCUUAACGUGCGCCUUUGGCGGUGGCCUCUCCGGG---CCGGGGAGC---------------------------------- ......(((((((((.((((((..((..............))..))))))))))))))).(((((((..---.))))))).---------------------------------- ( -31.74, z-score = -1.25, R) >droWil1.scaffold_180701 3069399 76 + 3904529 AAUUAAUCAUCGUCAUGGGGCGUGUGCACGAUUCGCUUAACGUGCGCCUUUGGCGGUGACUGCAAAGCAGGGAAAG--------------------------------------- ......(((((((((.((((((..((..............))..)))))))))))))))((((...))))......--------------------------------------- ( -27.54, z-score = -2.20, R) >droGri2.scaffold_15245 3470198 81 + 18325388 AAUUAAUCAUCGUCAUGGGGCGUGUGCACGAUUCGCUUAACGUGCGCCGCUGGCGGUGGUUUUCU-GCAUGUGGAGCCUAGC--------------------------------- ....(((((((((((....(((.(((((((..........)))))))))))))))))))))((((-(....)))))......--------------------------------- ( -27.80, z-score = -0.50, R) >droMoj3.scaffold_6496 10203942 82 - 26866924 AAUUAAUCAUCGUCAGGGGGCGUGUGCACGAUUCGCUUAACGUGCGCCGCUGGCGAUGGAGCCGAUGGAGCUGGAGUCAGCC--------------------------------- ......(((((.......((((.(((((((..........)))))))))))(((......)))))))).((((....)))).--------------------------------- ( -31.10, z-score = -0.89, R) >droVir3.scaffold_12875 10324726 78 + 20611582 AAUUAAUCAUCGUCAUGGGGCGUGUGCACGAUUCGCUUAACGUGCGCCUCUGGCGGUGGCUUCG----GCGUGGAGCUGGCC--------------------------------- ......(((((((((.((((((..((..............))..)))))))))))))))..(((----((.....)))))..--------------------------------- ( -30.64, z-score = -1.21, R) >consensus AAUUAAUCAUCGUCAUGGGGCGUGUGCACGAUUCGCUUAACGUGCGCCUUUGGCGGUGGUUCCCCCG_A___CCAGUCUCCC_________________________________ ......(((((((((.((((((..((..............))..)))))))))))))))........................................................ (-22.92 = -22.83 + -0.09)

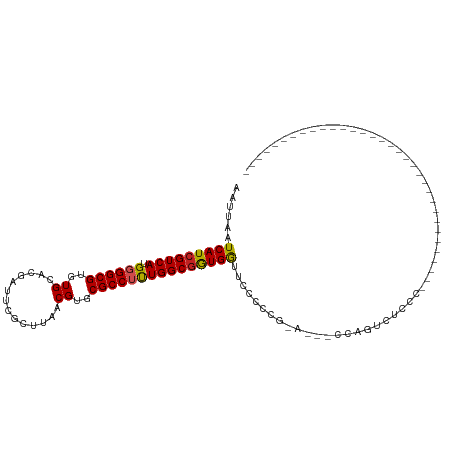
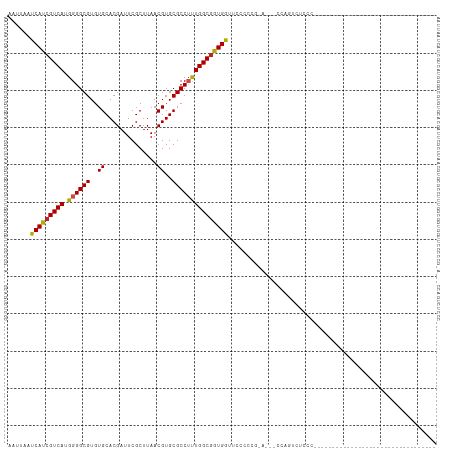
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:41:49 2011