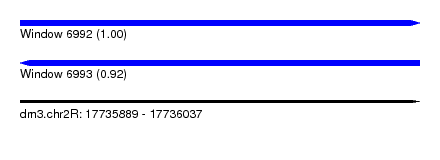
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,735,889 – 17,736,037 |
| Length | 148 |
| Max. P | 0.996787 |

| Location | 17,735,889 – 17,736,037 |
|---|---|
| Length | 148 |
| Sequences | 13 |
| Columns | 173 |
| Reading direction | forward |
| Mean pairwise identity | 71.18 |
| Shannon entropy | 0.59358 |
| G+C content | 0.49108 |
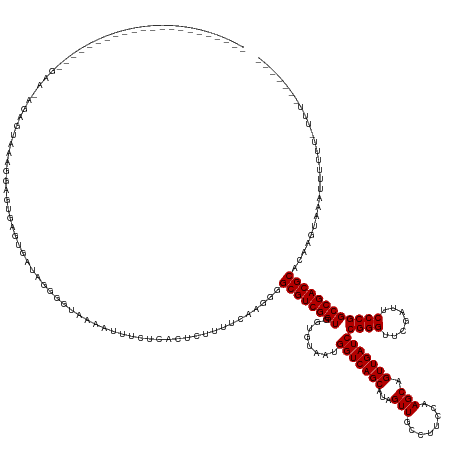
| Mean single sequence MFE | -38.74 |
| Consensus MFE | -28.98 |
| Energy contribution | -28.98 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.996787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
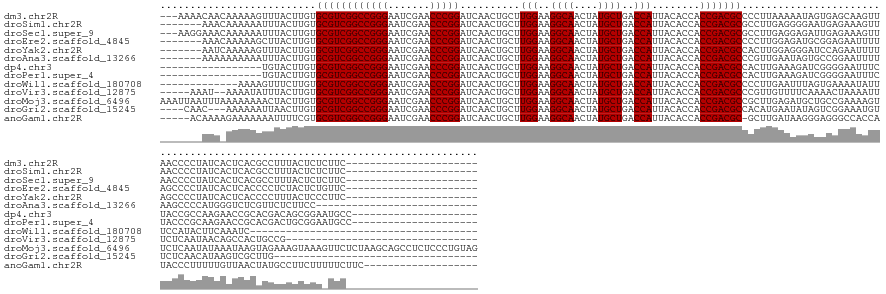
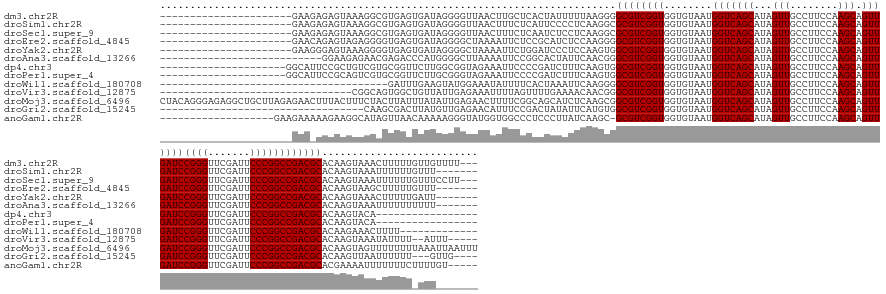
>dm3.chr2R 17735889 148 + 21146708 ---AAAACAACAAAAAGUUUACUUGUGCGUCGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGCCCCUUAAAAAUAGUGAGCAAGUUAACCCCUAUCACUCACGCCUUUACUCUCUUC---------------------- ---.............(((.((((((((((((((((((.......)))))..........(((..((((....))))..)))........)))))))..((.......))...)))))).)))............................---------------------- ( -34.80, z-score = -1.73, R) >droSim1.chr2R 16370385 144 + 19596830 -------AAACAAAAAAUUUACUUGUGCGUCGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGCGCCUUGAGGGGAAUGAGAAAGUUAACCCCUAUCACUCACGCCUUUACUCUCUUC---------------------- -------.................((((((((((((((.......)))))..........(((..((((....))))..)))........)))))))))...((((((..((((.................))))..)))...))).....---------------------- ( -40.33, z-score = -2.22, R) >droSec1.super_9 1051049 148 + 3197100 ---AAGGAAACAAAAAAUUUACUUGUGCGUCGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGCGCCUUGAGGAGAUUGAGAAAGUUAACCCCUAUCACUCACGCCUUUACUCUCUUC---------------------- ---..(....).............((((((((((((((.......)))))..........(((..((((....))))..)))........)))))))))...(((((((.((((...((...............))...)))).)))))))---------------------- ( -41.46, z-score = -2.42, R) >droEre2.scaffold_4845 11869239 144 + 22589142 -------AAACAAAAAGCUUACUUGUGCGUCGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGCCCCUUGGAGAUGCGGAGAAUUUUAGCCCCUAUCACUCACCCCUCUACUCUGUUC---------------------- -------.((((..............((((((((((((.......)))))..........(((..((((....))))..)))........)))))))....(((((..(..(((.....(((...)))...)))..).)))))...)))).---------------------- ( -34.90, z-score = -0.33, R) >droYak2.chr2R 9687871 144 - 21139217 -------AAUCAAAAAGUUUACUUGUGCGUCGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGCCACUUGGAGGGAUCCAGAAUUUUAGCCCCUAUCACUCACCCCUUUACUCCCUUC---------------------- -------................((.((((((((((((.......)))))..........(((..((((....))))..)))........)))))))))...(((((((...................................)))))))---------------------- ( -36.25, z-score = -0.81, R) >droAna3.scaffold_13266 11118524 139 + 19884421 -------AAAAAAAAAAUUUACUUGUGCGUCGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGCCCGUUGAAUAGUGCCGGAAUUUUAAGCCCCAUGGGUCUCGUUCUCUUCC--------------------------- -------.........(((((..((.((((((((((((.......)))))..........(((..((((....))))..)))........))))))).)).)))))......(((((.....((((...))))...))))).....--------------------------- ( -36.40, z-score = -0.21, R) >dp4.chr3 15290267 135 + 19779522 -----------------UGUACUUGUGCGUCGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGCCACUUGAAAGAUCGGGGAAUUUCUACCGCCAAGAACCGCACGACAGCGGAAUGCC--------------------- -----------------....((((.((((((((((((.......)))))..........(((..((((....))))..)))........)))))))..(((((....))))).............))))..((((......))))......--------------------- ( -41.60, z-score = -1.26, R) >droPer1.super_4 3401467 135 - 7162766 -----------------UGUACUUGUGCGUCGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGCCACUUGAAAGAUCGGGGAAUUUCUACCCGCAAGAACCGCACGACUGCGGAAUGCC--------------------- -----------------....(((((((((((((((((.......)))))..........(((..((((....))))..)))........))))))).............(((.........))))))))..(((((....)))))......--------------------- ( -46.00, z-score = -2.65, R) >droWil1.scaffold_180708 9630070 122 - 12563649 -------------AAAAGUUUCUUGUGCGUCGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGCCCCUUGAAUUUAGUGAAAAUAUUUCCAUACUUCAAAUC-------------------------------------- -------------..((((((...(.((((((((((((.......)))))..........(((..((((....))))..)))........))))))).)...))))))...........................-------------------------------------- ( -30.50, z-score = -1.42, R) >droVir3.scaffold_12875 10297286 134 + 20611582 -----AAAU--AAAAUAUUUACUUGUGCGUCGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGCCCGUUGUUUUCAAAACUAAAAUUUCUCAAUAACAGCCACUGCCG-------------------------------- -----....--.............(.((((((((((((.......)))))..........(((..((((....))))..)))........))))))).)((((((......................))))))........-------------------------------- ( -32.15, z-score = -1.79, R) >droMoj3.scaffold_6496 10165864 173 - 26866924 AAAUUAAUUUAAAAAAAACUACUUGUGCGUCGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGCCGCUUGAGAUGCUGCCGAAAAGUUCUCAAUAUAAAUAAGUAGAAAGUAAAGUUCUCUAAGCAGCCUCUCCCUGUAG ..................((((((((((((((((((((.......)))))..........(((..((((....))))..)))........)))))))...((((((.(((.......))))))))).....)))))))).................((((.......)))).. ( -46.40, z-score = -2.55, R) >droGri2.scaffold_15245 3443195 132 + 18325388 ----CAAC---AAAAAAUUAACUUGUGCGUCGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGCCACAUGAAUAUAGUCGGAAAUGUUCUCAACAUAAGUCGCUUG---------------------------------- ----....---.........((((((((((((((((((.......)))))..........(((..((((....))))..)))........))))))).....((((((........)))))).....))))))......---------------------------------- ( -35.20, z-score = -1.99, R) >anoGam1.chr2R 23158179 148 - 62725911 -----ACAAAAGAAAAAAAUUUUCGUGCGUCGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGC-GCUUGAUAAGGGAGGGCCACCAUACCCUUUUUGUUAACUAUGCCUUCUUUUUCUUC------------------- -----....((((((((.......((((((((((((((.......)))))..........(((..((((....))))..)))........)))))))-))(((((((((.((((........)))))))))))))..........)))))))).------------------- ( -47.60, z-score = -3.61, R) >consensus _______AAA_AAAAAAUUUACUUGUGCGUCGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGCCCCUUGAAAAGAGUGAGAAAUUUUACCCCUAUCACUCACGCCUUUACUCU_UUC______________________ ..........................((((((((((((.......)))))..........(((..((((....))))..)))........)))))))............................................................................ (-28.98 = -28.98 + 0.00)
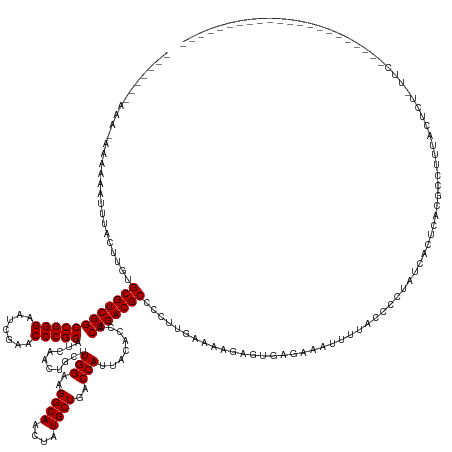
| Location | 17,735,889 – 17,736,037 |
|---|---|
| Length | 148 |
| Sequences | 13 |
| Columns | 173 |
| Reading direction | reverse |
| Mean pairwise identity | 71.18 |
| Shannon entropy | 0.59358 |
| G+C content | 0.49108 |
| Mean single sequence MFE | -43.08 |
| Consensus MFE | -30.05 |
| Energy contribution | -30.05 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.922436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17735889 148 - 21146708 ----------------------GAAGAGAGUAAAGGCGUGAGUGAUAGGGGUUAACUUGCUCACUAUUUUUAAGGGGCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCGACGCACAAGUAAACUUUUUGUUGUUUU--- ----------------------..(((((((....((.(((((((((((......)))).)))))...........((((((((........(((((((...(((........))).)))))))((((.......)))))))))))).)).))..)))))))........--- ( -43.50, z-score = -0.63, R) >droSim1.chr2R 16370385 144 - 19596830 ----------------------GAAGAGAGUAAAGGCGUGAGUGAUAGGGGUUAACUUUCUCAUUCCCCUCAAGGCGCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCGACGCACAAGUAAAUUUUUUGUUU------- ----------------------.((((((((..(((.(.((((((.((((.....)))).))))))))))....))((((((((........(((((((...(((........))).)))))))((((.......)))))))))))).........))))))....------- ( -44.30, z-score = -1.09, R) >droSec1.super_9 1051049 148 - 3197100 ----------------------GAAGAGAGUAAAGGCGUGAGUGAUAGGGGUUAACUUUCUCAAUCUCCUCAAGGCGCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCGACGCACAAGUAAAUUUUUUGUUUCCUU--- ----------------------((.((((......((....))((.((((.....)))).))..)))).))((((.((((((((........(((((((...(((........))).)))))))((((.......))))))))))))(((((.......)))))..))))--- ( -44.20, z-score = -0.86, R) >droEre2.scaffold_4845 11869239 144 - 22589142 ----------------------GAACAGAGUAGAGGGGUGAGUGAUAGGGGCUAAAAUUCUCCGCAUCUCCAAGGGGCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCGACGCACAAGUAAGCUUUUUGUUU------- ----------------------((((((((.((.((((...(((...((((........)))).))))))).....((((((((........(((((((...(((........))).)))))))((((.......)))))))))))).........))))))))))------- ( -44.70, z-score = -0.10, R) >droYak2.chr2R 9687871 144 + 21139217 ----------------------GAAGGGAGUAAAGGGGUGAGUGAUAGGGGCUAAAAUUCUGGAUCCCUCCAAGUGGCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCGACGCACAAGUAAACUUUUUGAUU------- ----------------------..(((((((...((((.((....(((((.......)))))..)).))))..(((.(((((((........(((((((...(((........))).)))))))((((.......))))))))))))))......)))))))....------- ( -42.90, z-score = 0.28, R) >droAna3.scaffold_13266 11118524 139 - 19884421 ---------------------------GGAAGAGAACGAGACCCAUGGGGCUUAAAAUUCCGGCACUAUUCAACGGGCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCGACGCACAAGUAAAUUUUUUUUUU------- ---------------------------(((((((((.....((....))((((......(((...........)))((((((((........(((((((...(((........))).)))))))((((.......))))))))))))..))))...))))))))).------- ( -40.00, z-score = 0.06, R) >dp4.chr3 15290267 135 - 19779522 ---------------------GGCAUUCCGCUGUCGUGCGGUUCUUGGCGGUAGAAAUUCCCCGAUCUUUCAAGUGGCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCGACGCACAAGUACA----------------- ---------------------((((......))))((((.....((((.((.(....).))))))........(((.(((((((........(((((((...(((........))).)))))))((((.......))))))))))))))..)))).----------------- ( -43.80, z-score = -0.08, R) >droPer1.super_4 3401467 135 + 7162766 ---------------------GGCAUUCCGCAGUCGUGCGGUUCUUGCGGGUAGAAAUUCCCCGAUCUUUCAAGUGGCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCGACGCACAAGUACA----------------- ---------------------.(((..(((((....)))))....)))(((.(....).)))...........(((.(((((((........(((((((...(((........))).)))))))((((.......)))))))))))))).......----------------- ( -45.50, z-score = -0.74, R) >droWil1.scaffold_180708 9630070 122 + 12563649 --------------------------------------GAUUUGAAGUAUGGAAAUAUUUUCACUAAAUUCAAGGGGCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCGACGCACAAGAAACUUUU------------- --------------------------------------..((((((...(((((....))))).....))))))..((((((((........(((((((...(((........))).)))))))((((.......)))))))))))).............------------- ( -34.40, z-score = -0.80, R) >droVir3.scaffold_12875 10297286 134 - 20611582 --------------------------------CGGCAGUGGCUGUUAUUGAGAAAUUUUAGUUUUGAAAACAACGGGCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCGACGCACAAGUAAAUAUUUU--AUUU----- --------------------------------..((.....(((((.((..(((((....)))))...)).)))))((((((((........(((((((...(((........))).)))))))((((.......))))))))))))....)).........--....----- ( -35.70, z-score = -0.18, R) >droMoj3.scaffold_6496 10165864 173 + 26866924 CUACAGGGAGAGGCUGCUUAGAGAACUUUACUUUCUACUUAUUUAUAUUGAGAACUUUUCGGCAGCAUCUCAAGCGGCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCGACGCACAAGUAGUUUUUUUUAAAUUAAUUU ((((...((((.((((((((((((.......))))))((((.......))))........)))))).)))).....((((((((........(((((((...(((........))).)))))))((((.......))))))))))))....)))).................. ( -53.10, z-score = -2.14, R) >droGri2.scaffold_15245 3443195 132 - 18325388 ----------------------------------CAAGCGACUUAUGUUGAGAACAUUUCCGACUAUAUUCAUGUGGCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCGACGCACAAGUUAAUUUUUU---GUUG---- ----------------------------------...((((((.(((((((..((((..(((((..(((....)))..)))))..)))).....))))))))))))).......((.((((..(((((.......))))).))))))(((((.......)))---))..---- ( -41.70, z-score = -2.21, R) >anoGam1.chr2R 23158179 148 + 62725911 -------------------GAAGAAAAAGAAGGCAUAGUUAACAAAAAGGGUAUGGUGGCCCUCCCUUAUCAAGC-GCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCGACGCACGAAAAUUUUUUUCUUUUGU----- -------------------(((((((((((..((...(....)...(((((...((....)).))))).....))-((((((((........(((((((...(((........))).)))))))((((.......)))))))))))).......)))))))))))...----- ( -46.20, z-score = -1.57, R) >consensus ______________________GAA_AGAGUAAAGGAGUGAGUGAUAGGGGUAAAAUUUCUCACUCUUUUCAAGGGGCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCGACGCACAAGUAAAUUUUUU_UUU_______ ............................................................................((((((((........(((((((...(((........))).)))))))((((.......)))))))))))).......................... (-30.05 = -30.05 + -0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:41:45 2011