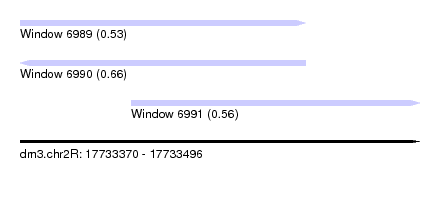
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,733,370 – 17,733,496 |
| Length | 126 |
| Max. P | 0.656969 |

| Location | 17,733,370 – 17,733,460 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 69.20 |
| Shannon entropy | 0.55745 |
| G+C content | 0.53838 |
| Mean single sequence MFE | -22.75 |
| Consensus MFE | -10.52 |
| Energy contribution | -10.83 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.533943 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 17733370 90 + 21146708 CCCCGUUGCAGGCACUACAGGCACU---ACUUGGCCUACAACUCCGUCUCUUUCGAAGUCUUCCGAUGGCCAUUCUAGUUGGUCA--UGACUAUG ....((((.((((....((((....---.)))))))).))))...(((....(((........)))((((((.......))))))--.))).... ( -21.90, z-score = -0.69, R) >droYak2.chr2R 9685158 86 - 21139217 CCCCGUUGCAGGCACUACU---------ACUUGGCCUCAAACUCCGUCUCUUUCGACGUCUUGUGUUGGCCAUUCUAGUUGGCUAGUUGGCCAUG .....(((.((((.(....---------....))))))))....((((......))))........((((((..((((....)))).)))))).. ( -25.30, z-score = -1.87, R) >droEre2.scaffold_4845 11866659 95 + 22589142 CCCCGUUGCUGGCACUACUUGGCCUCAAACUCCGUCUACGUCUCCGUCUCUUUCGACGUCUUGCGAUGGCCAUUCUAGUUGGCUAGAUGGCUAUG ....(((...(((.(.....))))...)))..(((..(((((............)))))...)))((((((((.((((....)))))))))))). ( -28.20, z-score = -2.25, R) >droSec1.super_9 1048397 81 + 3197100 CCCCGCUGCAGGCACUACU------------UGGCCUACAACUCCGUAUCUUUCGACGUCUUCCGAUAGCCAUUUUAGUUGGCCA--UGACUAUG ..........(((((((..------------((((.(((......)))....(((........)))..))))...))))..))).--........ ( -13.90, z-score = 0.32, R) >droSim1.chr2R 16367686 81 + 19596830 CCCCGUUGCAGGCACUACU------------UGGCCUACAACUCCGUCUCUUUCGACGUCUUCCGAUGGCCAUUCUAGUUGGCCA--UGACUAUG ....((((.((((.(....------------.))))).))))..((((......)))).......(((((((.......))))))--)....... ( -25.30, z-score = -3.36, R) >droWil1.scaffold_180701 3023632 78 + 3904529 CCCCGUUGGAAGCGCAA----------------ACCUGGUGCGUGUUUUGUUUGGCCUCUUUGGCUUGGUCGGUCGAAU-GGUCGGAUGGUCGAA ..(((((.((.(((((.----------------......)))))...(..(((((((.((.......))..))))))).-.))).)))))..... ( -21.90, z-score = 0.65, R) >consensus CCCCGUUGCAGGCACUACU____________UGGCCUACAACUCCGUCUCUUUCGACGUCUUCCGAUGGCCAUUCUAGUUGGCCA__UGACUAUG ....((((.((((....................)))).))))...............(((......((((((.......))))))...))).... (-10.52 = -10.83 + 0.31)



| Location | 17,733,370 – 17,733,460 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 69.20 |
| Shannon entropy | 0.55745 |
| G+C content | 0.53838 |
| Mean single sequence MFE | -25.34 |
| Consensus MFE | -9.84 |
| Energy contribution | -12.82 |
| Covariance contribution | 2.97 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.656969 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 17733370 90 - 21146708 CAUAGUCA--UGACCAACUAGAAUGGCCAUCGGAAGACUUCGAAAGAGACGGAGUUGUAGGCCAAGU---AGUGCCUGUAGUGCCUGCAACGGGG ........--......((((...(((((.((....))(((((.......))))).....)))))..)---))).(((((.((....)).))))). ( -24.70, z-score = -0.48, R) >droYak2.chr2R 9685158 86 + 21139217 CAUGGCCAACUAGCCAACUAGAAUGGCCAACACAAGACGUCGAAAGAGACGGAGUUUGAGGCCAAGU---------AGUAGUGCCUGCAACGGGG ..((((((.((((....))))..))))))...((((.((((......))))...))))...((..((---------.((((...)))).))..)) ( -25.60, z-score = -1.59, R) >droEre2.scaffold_4845 11866659 95 - 22589142 CAUAGCCAUCUAGCCAACUAGAAUGGCCAUCGCAAGACGUCGAAAGAGACGGAGACGUAGACGGAGUUUGAGGCCAAGUAGUGCCAGCAACGGGG .....((.(((((....))))).(((((.((....))((((.......(((....))).))))........))))).((.((....)).)))).. ( -28.20, z-score = -1.90, R) >droSec1.super_9 1048397 81 - 3197100 CAUAGUCA--UGGCCAACUAAAAUGGCUAUCGGAAGACGUCGAAAGAUACGGAGUUGUAGGCCA------------AGUAGUGCCUGCAGCGGGG ....((((--((((((.......))))))).....)))(((....))).....((((((((((.------------....).))))))))).... ( -28.10, z-score = -2.93, R) >droSim1.chr2R 16367686 81 - 19596830 CAUAGUCA--UGGCCAACUAGAAUGGCCAUCGGAAGACGUCGAAAGAGACGGAGUUGUAGGCCA------------AGUAGUGCCUGCAACGGGG ....(((.--((((((.......))))))((....))..((....)))))...((((((((((.------------....).))))))))).... ( -32.00, z-score = -4.08, R) >droWil1.scaffold_180701 3023632 78 - 3904529 UUCGACCAUCCGACC-AUUCGACCGACCAAGCCAAAGAGGCCAAACAAAACACGCACCAGGU----------------UUGCGCUUCCAACGGGG ........((((...-..((....))..........((((((((((..............))----------------))).)))))...)))). ( -13.44, z-score = 0.86, R) >consensus CAUAGCCA__UGGCCAACUAGAAUGGCCAUCGCAAGACGUCGAAAGAGACGGAGUUGUAGGCCA____________AGUAGUGCCUGCAACGGGG .....((....(((((.......))))).........((((......))))..(((((((((....................))))))))))).. ( -9.84 = -12.82 + 2.97)


| Location | 17,733,405 – 17,733,496 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 65.07 |
| Shannon entropy | 0.63443 |
| G+C content | 0.51366 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -11.29 |
| Energy contribution | -12.34 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.564911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17733405 91 + 21146708 ------------CAACUCCGUCUCUUUCGAAGUCU---UCCGAUGGCCAUUCUAGUUGGUCA--UGACUAUGUG-GCG---UAUGUGCGUGUUUCUGG--CGGUCACAUGCAAU ------------.......(((.....((.((((.---....(((((((.......))))))--))))).)).)-))(---((((((..(((.....)--))..)))))))... ( -23.20, z-score = -0.22, R) >droAna3.scaffold_13266 11115445 102 + 19884421 ------------GAACCCUGUCUUCGGCUGCUCCUCCAUUCGAUAGCCACACACUAUAUGCACCAACAUGUGUGUAUAUAUGGAGUGUGCGUGCUGGGGACCGUCGCAUGCAAU ------------.......((((((((((((.(((((((...(((..((((((...............))))))..))))))))).).))).)))))))))............. ( -31.66, z-score = -0.62, R) >droYak2.chr2R 9685187 96 - 21139217 ------------AAACUCCGUCUCUUUCGACGUCU---UGUGUUGGCCAUUCUAGUUGGCUAGUUGGCCAUGUG-GCGGCGUAUGUGCGUGUUUCUGG--CGGUCACAUGCAAU ------------..((.(((((.(....((((...---..))))(((((..((((....)))).)))))..).)-)))).))...(((((((..(...--..)..))))))).. ( -31.60, z-score = -1.48, R) >droEre2.scaffold_4845 11866685 105 + 22589142 AAACUCCGUCUACGUCUCCGUCUCUUUCGACGUCU---UGCGAUGGCCAUUCUAGUUGGCUAGAUGGCUAUGUG-GCG---UAUGUGCGUGUUUCUGG--CGGUCACAUGCAAU ..........((((((..((((......))))...---.((.((((((((.((((....)))))))))))))))-)))---))..(((((((..(...--..)..))))))).. ( -34.80, z-score = -2.03, R) >droSec1.super_9 1048423 91 + 3197100 ------------CAACUCCGUAUCUUUCGACGUCU---UCCGAUAGCCAUUUUAGUUGGCCA--UGACUAUGUG-GCG---UAUGUGCGUGUUCCUGG--CGGUCACAUGCAAU ------------......(((((...(((......---..)))..(((((..(((((.....--.))))).)))-)))---))))(((((((.((...--.))..))))))).. ( -24.00, z-score = -0.40, R) >droSim1.chr2R 16367712 91 + 19596830 ------------CAACUCCGUCUCUUUCGACGUCU---UCCGAUGGCCAUUCUAGUUGGCCA--UGACUAUGUG-GCG---UAUGUGCGUGUUCCUGG--CGGUCACAUGCAAU ------------..((.(((((......)))(((.---....(((((((.......))))))--)))).....)-).)---)...(((((((.((...--.))..))))))).. ( -27.50, z-score = -1.06, R) >droWil1.scaffold_180701 3023654 82 + 3904529 -------------GUGCGUGUUUUGUUUGGCCUCU---U----UGGC----UUGGUCGGUCGAAUGGUCGGAUG-GUC------GAAUGGUUAGCUGGU-CGGUCACAUGCAAU -------------.(((((((...((((((((..(---(----((((----(.....))))))).))))))))(-..(------((..((....))..)-))..)))))))).. ( -29.90, z-score = -2.98, R) >consensus ____________CAACUCCGUCUCUUUCGACGUCU___UCCGAUGGCCAUUCUAGUUGGCCA__UGACUAUGUG_GCG___UAUGUGCGUGUUCCUGG__CGGUCACAUGCAAU ..................((((......))))...........((((((.......)))))).......................(((((((..(((...)))..))))))).. (-11.29 = -12.34 + 1.05)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:41:43 2011