| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,659,551 – 17,659,660 |
| Length | 109 |
| Max. P | 0.733964 |

| Location | 17,659,551 – 17,659,660 |
|---|---|
| Length | 109 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 70.45 |
| Shannon entropy | 0.57855 |
| G+C content | 0.48142 |
| Mean single sequence MFE | -32.57 |
| Consensus MFE | -11.90 |
| Energy contribution | -10.96 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.682012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
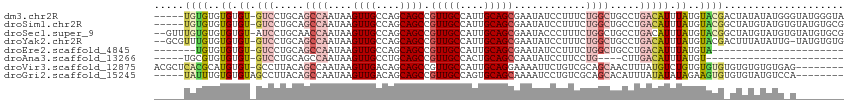
>dm3.chr2R 17659551 109 + 21146708 -----UGUGUGUGUGU-GUCCUGCAGCCAAUAAGUUGCCAGCAGCCGUUGCCAUUGCAGCGAAUAUCCUUUCUGGCUGCCUGACAUUUAUGUACGACUAUAUAUGGGUAUGGGUA -----.(((..((.((-(((..(((((((....((((....))))((((((....))))))...........)))))))..))))).))..)))..((((((....))))))... ( -39.50, z-score = -2.09, R) >droSim1.chr2R 16300655 109 + 19596830 -----UGUGUGUGUGU-GUCCUGCAGCCAAUAAGUUGCCAGCAGCCGUUGCCAUUGCAGCGAAUAUCCUUUCUGGCUGCCUGACAUUUAUGUACGGCUAUGUAUGUGUAUGUGCG -----.(((..((.((-(((..(((((((....((((....))))((((((....))))))...........)))))))..))))).))..))).((.(((((....))))))). ( -38.00, z-score = -1.92, R) >droSec1.super_9 981875 112 + 3197100 --GUUUGUGUGUGUGU-AUCCUGCAACCAAUAAGUUGCCAGCAGCCGUUGCCAUUGCAGCGAAUACCCUUUCUGGCUGCCUGACAUUUAUGUACGGCUAUGUAUGUGUAUGUGCG --((.((..((((..(-(.((.(((((......)))))(((((((((((((....)))))(((......))).))))).)))............)).))..))))..))...)). ( -33.70, z-score = -0.98, R) >droYak2.chr2R 9618877 111 - 21139217 --GCGUUUGUGUGUGU-GUCCUGCAGCCAAUAAGUUGCCAGCAGCCGUUGCCAUUGCAGCGAAUAUCCUUUCUGGCUGCCUGACAUUUAUGUACGACUUUAUAUUG-UAUGUGUG --....(((((..(((-(((..(((((((....((((....))))((((((....))))))...........)))))))..)))))..)..)))))..........-........ ( -33.90, z-score = -1.98, R) >droEre2.scaffold_4845 11799783 85 + 22589142 -------UGUGUGUGU-GUCCUGCAGCCAAUAAGUUGCCAGCAGCCGUUGCCAUUGCAGCGAAUAUCCUUUCUGGCUGCCUGACAUUUAUGUA---------------------- -------((..((.((-(((..(((((((....((((....))))((((((....))))))...........)))))))..))))).))..))---------------------- ( -31.40, z-score = -3.47, R) >droAna3.scaffold_13266 11045943 82 + 19884421 -----UGCGUGUGUGU-GUCCUGCAGCCAAUAAGUUGCCUGCAGCCGUUGCCACUGCAGCCAAUAUCCUUCCUG----CUUGACAUUUAUGU----------------------- -----.(((((...((-(((..((((.......((((.((((((.........)))))).)))).......)))----)..))))).)))))----------------------- ( -25.44, z-score = -2.29, R) >droVir3.scaffold_12875 10210319 106 + 20611582 ACGCUCACGCAUGUGU-GCCUUACAGCCAAUAAGUUGACAGCAGCCGUUGCCAUUGCAGGAAAAUUCUGUCGCAGCAACUUUAUGUCUGUGUGUGUGUGUGUGUGAG-------- ...((((((((((..(-((..(((((.((....((((....)))).(((((....((((((...))))))....)))))....)).))))).)))..))))))))))-------- ( -35.70, z-score = -1.13, R) >droGri2.scaffold_15245 3367168 102 + 18325388 -----UAUUUGUGUGUAGCCUUACAGCCAAUAAGUUGACAGCAGCCGUUGCCAGUGCAGCAAAAUCCUGUCGCAGCACAUUUAUAUAUAGAAGUGUGUGUAUGUCCA-------- -----.....((((((.((((((.......))))..(((((.....(((((....)))))......))))))).)))))).((((((((....))))))))......-------- ( -22.90, z-score = 0.02, R) >consensus _____UGUGUGUGUGU_GUCCUGCAGCCAAUAAGUUGCCAGCAGCCGUUGCCAUUGCAGCGAAUAUCCUUUCUGGCUGCCUGACAUUUAUGUACG_CUAUAUAUG_G________ .......((((.((((.((...(((((......)))))..))....(((((....)))))......................)))).))))........................ (-11.90 = -10.96 + -0.94)

| Location | 17,659,551 – 17,659,660 |
|---|---|
| Length | 109 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 70.45 |
| Shannon entropy | 0.57855 |
| G+C content | 0.48142 |
| Mean single sequence MFE | -28.26 |
| Consensus MFE | -12.20 |
| Energy contribution | -13.30 |
| Covariance contribution | 1.10 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.733964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
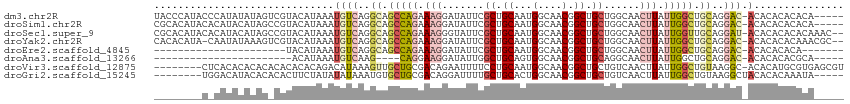
>dm3.chr2R 17659551 109 - 21146708 UACCCAUACCCAUAUAUAGUCGUACAUAAAUGUCAGGCAGCCAGAAAGGAUAUUCGCUGCAAUGGCAACGGCUGCUGGCAACUUAUUGGCUGCAGGAC-ACACACACACA----- ..............................((((..((((((((.((((.....)((((((.((....))..))).)))..))).))))))))..)))-)..........----- ( -30.60, z-score = -1.33, R) >droSim1.chr2R 16300655 109 - 19596830 CGCACAUACACAUACAUAGCCGUACAUAAAUGUCAGGCAGCCAGAAAGGAUAUUCGCUGCAAUGGCAACGGCUGCUGGCAACUUAUUGGCUGCAGGAC-ACACACACACA----- ..............................((((..((((((((.((((.....)((((((.((....))..))).)))..))).))))))))..)))-)..........----- ( -30.60, z-score = -1.57, R) >droSec1.super_9 981875 112 - 3197100 CGCACAUACACAUACAUAGCCGUACAUAAAUGUCAGGCAGCCAGAAAGGGUAUUCGCUGCAAUGGCAACGGCUGCUGGCAACUUAUUGGUUGCAGGAU-ACACACACACAAAC-- ..............(((((((((.......(((((.(((((..(((......))))))))..)))))))))))).))((((((....)))))).....-..............-- ( -30.01, z-score = -1.31, R) >droYak2.chr2R 9618877 111 + 21139217 CACACAUA-CAAUAUAAAGUCGUACAUAAAUGUCAGGCAGCCAGAAAGGAUAUUCGCUGCAAUGGCAACGGCUGCUGGCAACUUAUUGGCUGCAGGAC-ACACACACAAACGC-- ........-.....................((((..((((((((.((((.....)((((((.((....))..))).)))..))).))))))))..)))-).............-- ( -30.60, z-score = -2.35, R) >droEre2.scaffold_4845 11799783 85 - 22589142 ----------------------UACAUAAAUGUCAGGCAGCCAGAAAGGAUAUUCGCUGCAAUGGCAACGGCUGCUGGCAACUUAUUGGCUGCAGGAC-ACACACACA------- ----------------------........((((..((((((((.((((.....)((((((.((....))..))).)))..))).))))))))..)))-)........------- ( -30.60, z-score = -3.61, R) >droAna3.scaffold_13266 11045943 82 - 19884421 -----------------------ACAUAAAUGUCAAG----CAGGAAGGAUAUUGGCUGCAGUGGCAACGGCUGCAGGCAACUUAUUGGCUGCAGGAC-ACACACACGCA----- -----------------------.......((((..(----(((..((....(((.((((((((....).))))))).)))....))..))))..)))-)..........----- ( -27.90, z-score = -2.74, R) >droVir3.scaffold_12875 10210319 106 - 20611582 --------CUCACACACACACACACACAGACAUAAAGUUGCUGCGACAGAAUUUUCCUGCAAUGGCAACGGCUGCUGUCAACUUAUUGGCUGUAAGGC-ACACAUGCGUGAGCGU --------(((((............((((.((..(((((((.(((.(((.......))))...(((....))))).).))))))..)).))))...((-(....))))))))... ( -25.20, z-score = 0.77, R) >droGri2.scaffold_15245 3367168 102 - 18325388 --------UGGACAUACACACACUUCUAUAUAUAAAUGUGCUGCGACAGGAUUUUGCUGCACUGGCAACGGCUGCUGUCAACUUAUUGGCUGUAAGGCUACACACAAAUA----- --------............................((((..(((((((.(..(((((.....)))))....).)))))..(((((.....)))))))..))))......----- ( -20.60, z-score = 0.41, R) >consensus ________C_CAUACAUAG_CGUACAUAAAUGUCAGGCAGCCAGAAAGGAUAUUCGCUGCAAUGGCAACGGCUGCUGGCAACUUAUUGGCUGCAGGAC_ACACACACACA_____ ...............................(((..((.(((((.(((.......((.((...(....).)).))......))).))))).))..)))................. (-12.20 = -13.30 + 1.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:41:36 2011