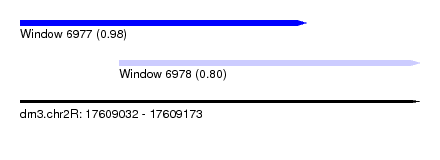
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,609,032 – 17,609,173 |
| Length | 141 |
| Max. P | 0.979498 |

| Location | 17,609,032 – 17,609,133 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 68.14 |
| Shannon entropy | 0.59181 |
| G+C content | 0.41139 |
| Mean single sequence MFE | -22.58 |
| Consensus MFE | -11.77 |
| Energy contribution | -11.28 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17609032 101 + 21146708 UAUUACAAAUUUCGAGCUUCUGUUUCUGUGUGUUUUAUGAAGCUGAUAAGCAGC------ACCCACCGCCCUGCUUA-UCAGCUUAGAGUUCUGCCCUCAUAUGCUAC .............((((....))))..((((((......((((((((((((((.------.(.....)..)))))))-))))))).(((.......)))))))))... ( -25.60, z-score = -2.47, R) >droSim1.chr2R 16260572 107 + 19596830 UAUUACAACUUUCGAGCUUCUGUUUCUGUGUGUUUUAUGGAACUGAUAAGCGGUUCCAUAACCCACCGCCCUGCUUA-UCAGUUUAGAGUUCUGCCUUCAUAUGCUAC .............(((((.(((...(((.(((..(((((((((((.....)))))))))))..))).((...))...-.)))..))))))))................ ( -26.60, z-score = -2.70, R) >droSec1.super_9 934941 101 + 3197100 UAUUACAAAUUUCGAACUUCUUUUUCUGUGUGUUUUAUGGAGCUAAUAAGCAGC------ACCCACCGCCCUGCUUA-UCAGUUUAGAGUUCUGCCUUCAUAUGCUAC .............((((((.....((((((.....))))))(((.((((((((.------.(.....)..)))))))-).)))...))))))................ ( -18.40, z-score = -0.78, R) >droYak2.chr2R 9577923 96 - 21139217 UAUUACAAAUUUCGAGCUUCUUUUUU--U---UUUUAUGAAGCUGAUAAGUAGC------ACCCCCCGCCCUGCUUA-UCAGUUUAGGGCUCUGCCCUCAUGUGUCAG ...((((...................--.---.......((((((((((((((.------.(.....)..)))))))-)))))))(((((...)))))..)))).... ( -24.30, z-score = -3.06, R) >droEre2.scaffold_4845 11758271 97 + 22589142 UAUUACAAAUGUCGAGGUUUUUCUUUGGU---UUGUUUGGUGCUGAUAAGUAGC------ACCCCC-GCCCUGCUUA-UCAGUUUAGAGUUCUGCCCUCAUGUGUUAU ....(((.(((..(.(((....((((((.---.........((((((((((((.------.(....-)..)))))))-)))))))))))....)))).))).)))... ( -21.60, z-score = -1.20, R) >droMoj3.scaffold_6540 18928103 101 - 34148556 UUCAAGGACUUUUAAGCUUACGAACUGUUGUUAUGCAUGCAGUAAAGGAAUACUCUC--AGUAAAGUAUCCUAAUUAAUUCAUUUAAAGUCCCGCUGUGCUGC----- .....(((((((...........(((((((.....)).)))))..((((.((((...--.....)))))))).............))))))).((......))----- ( -19.00, z-score = -0.30, R) >consensus UAUUACAAAUUUCGAGCUUCUGUUUCUGUGUGUUUUAUGGAGCUGAUAAGCAGC______ACCCACCGCCCUGCUUA_UCAGUUUAGAGUUCUGCCCUCAUAUGCUAC .............((((((....................((((((((((((((.................))))))).))))))).))))))................ (-11.77 = -11.28 + -0.49)

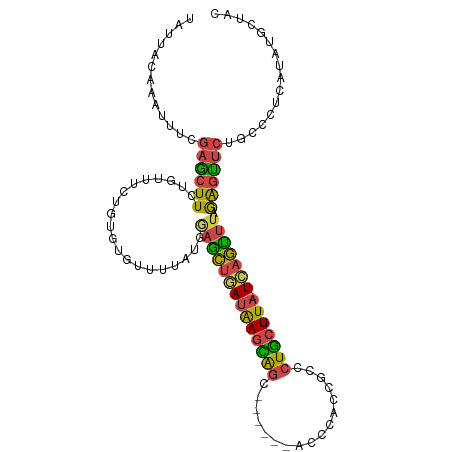
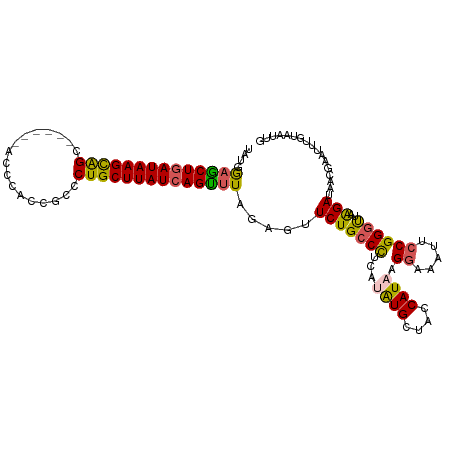
| Location | 17,609,067 – 17,609,173 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.98 |
| Shannon entropy | 0.34527 |
| G+C content | 0.43514 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -22.29 |
| Energy contribution | -21.73 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.803753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17609067 106 + 21146708 UAUGAAGCUGAUAAGCAGC------ACCCACCGCCCUGCUUAUCAGCUUAGAGUUCUGCCCUCAUAUGCUACCAUAAGGAAAUUCCGGGCUAAGAUAACGGAUUUGUAAUUG ....((((((((((((((.------.(.....)..)))))))))))))).((((((.((((...((((....)))).((.....))))))...(....)))))))....... ( -31.80, z-score = -2.84, R) >droSim1.chr2R 16260607 112 + 19596830 UAUGGAACUGAUAAGCGGUUCCAUAACCCACCGCCCUGCUUAUCAGUUUAGAGUUCUGCCUUCAUAUGCUACCAUUAGGAAAUUCCGGGUUAAGAUAACGAAUUUGUAAUUG ....((((((((((((((..(...........)..)))))))))))))).((((((.((........)).(((....((.....)).))).........))))))....... ( -26.00, z-score = -1.23, R) >droSec1.super_9 934976 106 + 3197100 UAUGGAGCUAAUAAGCAGC------ACCCACCGCCCUGCUUAUCAGUUUAGAGUUCUGCCUUCAUAUGCUACCAUUAGGAAAUUCCGGGUUAAGAUAAGGAAUUUGUAAUUG ....(((((.((((((((.------.(.....)..)))))))).))))).(((((((....((.((....(((....((.....)).))))).))...)))))))....... ( -21.80, z-score = 0.31, R) >droYak2.chr2R 9577953 106 - 21139217 UAUGAAGCUGAUAAGUAGC------ACCCCCCGCCCUGCUUAUCAGUUUAGGGCUCUGCCCUCAUGUGUCAGCAUAAGGAAAUUCCGGGUGAAGAUUACGGAUUUGUAAUUA ((((((((((((((((((.------.(.....)..))))))))))))))(((((...)))))))))..(((.(....((.....)).).))).(((((((....))))))). ( -31.40, z-score = -1.61, R) >droEre2.scaffold_4845 11758303 92 + 22589142 UUUGGUGCUGAUAAGUAGC------ACCC-CCGCCCUGCUUAUCAGUUUAGAGUUCUGCCCUCAUGUGUUAUCAUAAGGAAAUUACGGAUUUGGAAUUG------------- ......((((((((((((.------.(..-..)..))))))))))))....((((((..(((.(((......))).)))(((((...))))))))))).------------- ( -21.00, z-score = -0.64, R) >consensus UAUGGAGCUGAUAAGCAGC______ACCCACCGCCCUGCUUAUCAGUUUAGAGUUCUGCCCUCAUAUGCUACCAUAAGGAAAUUCCGGGUUAAGAUAACGAAUUUGUAAUUG ....((((((((((((((.................)))))))))))))).....(((((((...((((....)))).((.....))))))..)))................. (-22.29 = -21.73 + -0.56)

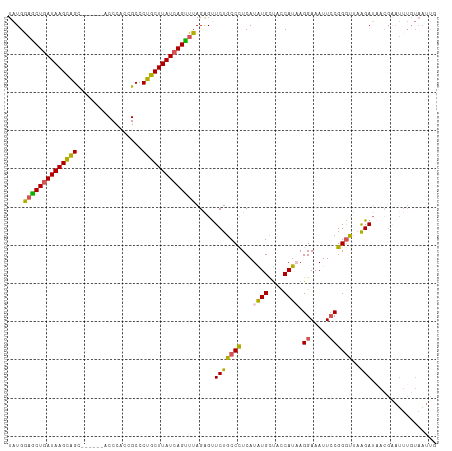
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:41:33 2011