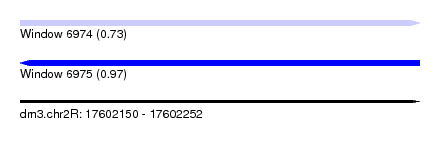
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,602,150 – 17,602,252 |
| Length | 102 |
| Max. P | 0.968828 |

| Location | 17,602,150 – 17,602,252 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 69.72 |
| Shannon entropy | 0.58928 |
| G+C content | 0.44744 |
| Mean single sequence MFE | -27.63 |
| Consensus MFE | -16.95 |
| Energy contribution | -17.16 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.22 |
| Mean z-score | -0.64 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.725488 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
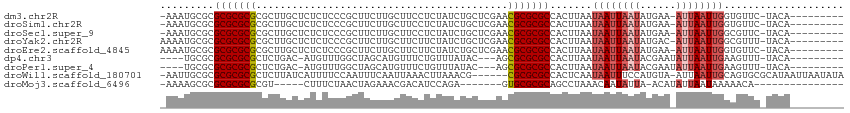
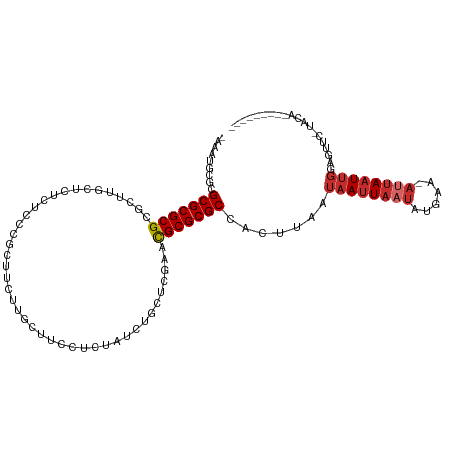
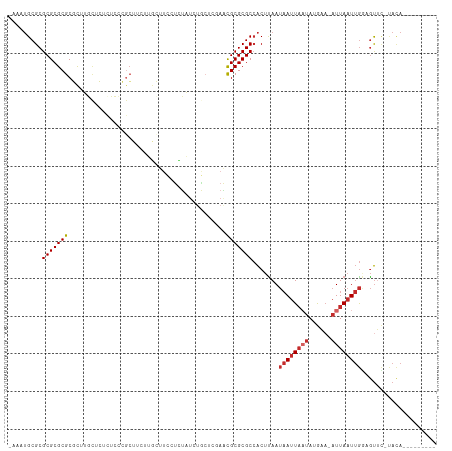
>dm3.chr2R 17602150 102 + 21146708 ---------UGUA-GAACACCAAUUAAU-UUCAUAUUAAUUAUUAAGUGGCGCGCGUUCGAGCAGAUAGAGGAAGCAAGAAGCGGGAGAGAGCAAGCGCGCGCGCGCGCAUUU- ---------....-...(((....((((-(.......)))))....)))((((((((.((.((.......(....).....((........))..)).)).))))))))....- ( -27.70, z-score = -0.60, R) >droSim1.chr2R 16253833 102 + 19596830 ---------UGUA-GAACACCAAUUAAU-UUCAUAUUAAUUAUUAAGUGGCGCGCGUUCGAGCAGAUAGAGGAAGCAAGAAGCGGGAGAGAGCAAGCGCGCGCGCGCGCAUUU- ---------....-...(((....((((-(.......)))))....)))((((((((.((.((.......(....).....((........))..)).)).))))))))....- ( -27.70, z-score = -0.60, R) >droSec1.super_9 928191 102 + 3197100 ---------UGUA-GAACGCCAAUUAAU-UUCAUAUUAAUUAUUAAGUGGCGCGCGUUCGAGCAGAUAGAGGAAGCAAGAAGCGGGAGAGAGCAAGCGCGCGCGCGCGCAUUU- ---------....-...(((....((((-(.......)))))....)))((((((((.((.((.......(....).....((........))..)).)).))))))))....- ( -27.30, z-score = 0.33, R) >droYak2.chr2R 9570895 103 - 21139217 ---------UGUA-AAACGCCAAUUAAU-GUCAUAUUAAUUAUUAAGUGGCGCGCGUUCGAGCAGAUAGAAGAAGCAAGAAGCGGGAGAGAGCAAGCGCGCGCGCGCGCAUUUU ---------(((.-...(((((.(((((-(..........)))))).)))))(((.(((..((...........))..)))))).......))).(((((....)))))..... ( -28.20, z-score = 0.16, R) >droEre2.scaffold_4845 11751278 103 + 22589142 ---------UGUA-GAACACCAAUUAAU-UUCAUAUUAAUUAUUAAGUGGCGCGCGUUCGAGCAGAUAGAAGAAGCAAGAAGCGGGAGAGAGCAAGCGCGCGCGCGCGCAUUUU ---------....-...(((....((((-(.......)))))....)))((((((((.((.((.....(......).....((........))..)).)).))))))))..... ( -26.30, z-score = -0.14, R) >dp4.chr3 15168906 96 + 19779522 ---------UGUA-AAACUUCAAUUAAUAUUCGUAUUAAUUAUUAAGUGGCGCGCGCU---GUAUAAACAGAAACAUGCUAGCCAAACAU-GUCAGAGCGCGCGCGCGCA---- ---------....-.......((((((((....)))))))).....((((((((((((---((....))....(((((.........)))-))...))))))))).))).---- ( -28.10, z-score = -1.41, R) >droPer1.super_4 3280618 96 - 7162766 ---------UGUA-AAACUUCAAUUAAUAUUCGUAUUAAUUAUUAAGUGGCGCGCGCU---GUAUAAACAGAAACAUGCUAGCCAAACAU-GUCAGAGCGCGCGCGCGCA---- ---------....-.......((((((((....)))))))).....((((((((((((---((....))....(((((.........)))-))...))))))))).))).---- ( -28.10, z-score = -1.41, R) >droWil1.scaffold_180701 2867222 106 + 3904529 UAUAUUAAUUAUGCGCACUGCAAUUAAU-UACAUGGAAAUUAUUGAGUGGCGCGCG------CGUUUAAGUUUAAUUGAAAUUGGAAAAUGAUAAGAGCGCGCGCGCGCAAUU- ...........(((.((((.((((.(((-(.......))))))))))))(((((((------(((((........(((..(((....)))..))))))))))))))))))...- ( -32.20, z-score = -1.87, R) >droMoj3.scaffold_6496 10003010 85 - 26866924 ---------------UGUUUUUAUUAAUAUGU-UAAUAUUGUUUAGGCUGCGCGCAC-------UCUGGAUGUCGUUUCUAGUUAGAAAG-----ACGCGCGCGCGCGCUUUU- ---------------.........((((((..-..))))))...(((((((((((.(-------....)..(((.(((((....))))))-----))..))))))).))))..- ( -23.10, z-score = -0.19, R) >consensus _________UGUA_GAACUCCAAUUAAU_UUCAUAUUAAUUAUUAAGUGGCGCGCGUUCGAGCAGAUAGAGGAAGCAAGAAGCGGGAGAGAGCAAGCGCGCGCGCGCGCAUUU_ .....................(((((((......))))))).....(((((((((((........................................)))))))).)))..... (-16.95 = -17.16 + 0.20)

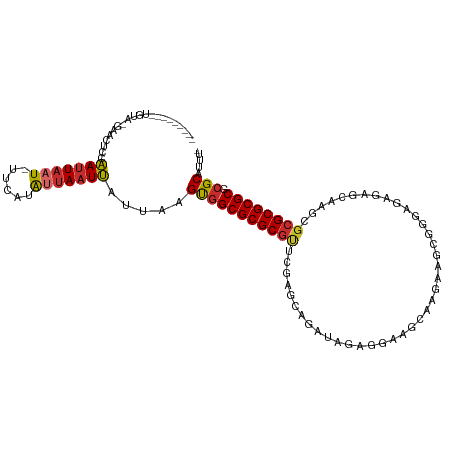
| Location | 17,602,150 – 17,602,252 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 69.72 |
| Shannon entropy | 0.58928 |
| G+C content | 0.44744 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -12.54 |
| Energy contribution | -13.22 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.968828 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2R 17602150 102 - 21146708 -AAAUGCGCGCGCGCGCGCUUGCUCUCUCCCGCUUCUUGCUUCCUCUAUCUGCUCGAACGCGCGCCACUUAAUAAUUAAUAUGAA-AUUAAUUGGUGUUC-UACA--------- -....(((((((..((.((............((.....))...........)).))..)))))))((((....(((((((.....-)))))))))))...-....--------- ( -24.50, z-score = -1.38, R) >droSim1.chr2R 16253833 102 - 19596830 -AAAUGCGCGCGCGCGCGCUUGCUCUCUCCCGCUUCUUGCUUCCUCUAUCUGCUCGAACGCGCGCCACUUAAUAAUUAAUAUGAA-AUUAAUUGGUGUUC-UACA--------- -....(((((((..((.((............((.....))...........)).))..)))))))((((....(((((((.....-)))))))))))...-....--------- ( -24.50, z-score = -1.38, R) >droSec1.super_9 928191 102 - 3197100 -AAAUGCGCGCGCGCGCGCUUGCUCUCUCCCGCUUCUUGCUUCCUCUAUCUGCUCGAACGCGCGCCACUUAAUAAUUAAUAUGAA-AUUAAUUGGCGUUC-UACA--------- -....(((((((((((..(..((........((.....))...........))..)..))))))).......((((((((.....-))))))))))))..-....--------- ( -25.21, z-score = -1.18, R) >droYak2.chr2R 9570895 103 + 21139217 AAAAUGCGCGCGCGCGCGCUUGCUCUCUCCCGCUUCUUGCUUCUUCUAUCUGCUCGAACGCGCGCCACUUAAUAAUUAAUAUGAC-AUUAAUUGGCGUUU-UACA--------- ((((((((((((((..((...((........((.....))...........)).))..))))))).......((((((((.....-))))))))))))))-)...--------- ( -26.91, z-score = -1.52, R) >droEre2.scaffold_4845 11751278 103 - 22589142 AAAAUGCGCGCGCGCGCGCUUGCUCUCUCCCGCUUCUUGCUUCUUCUAUCUGCUCGAACGCGCGCCACUUAAUAAUUAAUAUGAA-AUUAAUUGGUGUUC-UACA--------- .....(((((((..((.((............((.....))...........)).))..)))))))((((....(((((((.....-)))))))))))...-....--------- ( -24.50, z-score = -1.24, R) >dp4.chr3 15168906 96 - 19779522 ----UGCGCGCGCGCGCUCUGAC-AUGUUUGGCUAGCAUGUUUCUGUUUAUAC---AGCGCGCGCCACUUAAUAAUUAAUACGAAUAUUAAUUGAAGUUU-UACA--------- ----.(.((((((((.....(((-(((((.....))))))))..(((....))---))))))))))((((..(((((((((....)))))))))))))..-....--------- ( -30.40, z-score = -2.07, R) >droPer1.super_4 3280618 96 + 7162766 ----UGCGCGCGCGCGCUCUGAC-AUGUUUGGCUAGCAUGUUUCUGUUUAUAC---AGCGCGCGCCACUUAAUAAUUAAUACGAAUAUUAAUUGAAGUUU-UACA--------- ----.(.((((((((.....(((-(((((.....))))))))..(((....))---))))))))))((((..(((((((((....)))))))))))))..-....--------- ( -30.40, z-score = -2.07, R) >droWil1.scaffold_180701 2867222 106 - 3904529 -AAUUGCGCGCGCGCGCUCUUAUCAUUUUCCAAUUUCAAUUAAACUUAAACG------CGCGCGCCACUCAAUAAUUUCCAUGUA-AUUAAUUGCAGUGCGCAUAAUUAAUAUA -...(((((((((((((..(((........................)))..)------))))))).(((((((((((.......)-))).)))).))))))))........... ( -27.36, z-score = -2.89, R) >droMoj3.scaffold_6496 10003010 85 + 26866924 -AAAAGCGCGCGCGCGCGU-----CUUUCUAACUAGAAACGACAUCCAGA-------GUGCGCGCAGCCUAAACAAUAUUA-ACAUAUUAAUAAAAACA--------------- -...((.((((((((((((-----((((((....))))).))).(....)-------)))))))).))))......(((((-(....))))))......--------------- ( -23.20, z-score = -2.69, R) >consensus _AAAUGCGCGCGCGCGCGCUUGCUCUCUCCCGCUUCUUGCUUCCUCUAUCUGCUCGAACGCGCGCCACUUAAUAAUUAAUAUGAA_AUUAAUUGGAGUUC_UACA_________ .........(((((((..........................................))))))).......((((((((......)))))))).................... (-12.54 = -13.22 + 0.68)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:41:30 2011