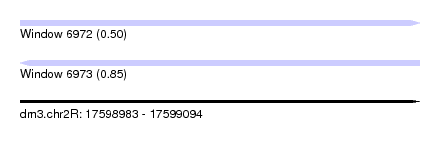
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,598,983 – 17,599,094 |
| Length | 111 |
| Max. P | 0.850231 |

| Location | 17,598,983 – 17,599,094 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.30 |
| Shannon entropy | 0.41823 |
| G+C content | 0.39211 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -11.93 |
| Energy contribution | -12.60 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
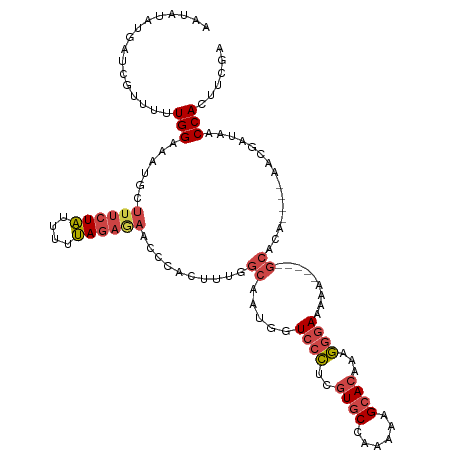
>dm3.chr2R 17598983 111 + 21146708 AAAAUAUAAUCGUUUUUGGAAAUGCUUUCUAUUUUUAGAGAACCCGCUUUGGCAAUGGUCCCUCGUGCCAAAAAGCACAAAGGGAAAAA-----GCACA----AACGAUAACCACUUCGA ........(((((((.......(((((((((((.((((((......)))))).)))))(((((.((((......))))..))))).)))-----))).)----))))))........... ( -25.70, z-score = -1.58, R) >droAna3.scaffold_13266 10997236 105 + 19884421 ------UUAACUGUGUUGGA----CAUCCUGUCCUCAAAUACACCUCU-----ACUGGUGCUAUCUUCAAAAAAGCACAAAACGAAAAAAUCUGAUAUAGCCCAGCUAUAACCACUAUGG ------.....(((((((((----(.....)))).......((((...-----...)))).............))))))................((((((...)))))).......... ( -20.30, z-score = -2.61, R) >droEre2.scaffold_4845 11747957 110 + 22589142 AAUAAAUGAUAUUUUUUGGUAGUGCUUUCUAUUUUUAGAAAACCCACUUAGGCAAUGGUCCCUCGUGCCAAAAAGCACAAAGGGAAAA------GCACA----AACGAUAACCACGUCGA .....................((((((((((....)))))).................(((((.((((......))))..)))))...------)))).----..((((......)))). ( -24.40, z-score = -1.32, R) >droYak2.chr2R 9567500 110 - 21139217 AAUAAAUGAUCUUUUUUGGUAGUGCUUUCUAUUUUUAGAGAACCCGCUUUGGCAAUGGUCCCACGUGCCAAAAAGCACAAAGGGAAAA------GCACA----AACGAUAACCACGCCGA ...............(((((.((((((((((((.((((((......)))))).)))))((((..((((......))))...)))))))------)))).----...(.....)..))))) ( -26.80, z-score = -1.22, R) >droSec1.super_9 924725 111 + 3197100 AAUAUAUGAUCGUUUUUGGAAAUGCUUUCUAUUUUUAGAGAACCCACUUUGGCAUUGGUCCCUCGUGCCAAAAAGCACUAAGGGAAAAA-----GCACA----AACGAUAACCACUUCGA ........(((((((.((..(((((((((((....)))))..........))))))..(((((.((((......))))..)))))....-----.)).)----))))))........... ( -25.50, z-score = -1.65, R) >droSim1.chr2R 16250334 111 + 19596830 AAUAUAUGAUCGUUUUUGGAAAUGUUUUCUAUUUCUAGAGAACCCACUUUGGCAUUGGUCCCUCGUGCCAAAAAGCACUAAGGGAAAAA-----GCACA----AACGAUAACCACUUCGA ........(((((((.(((....((((((((....))))))))))).....((.((..(((((.((((......))))..)))))..))-----))..)----))))))........... ( -28.20, z-score = -2.45, R) >consensus AAUAUAUGAUCGUUUUUGGAAAUGCUUUCUAUUUUUAGAGAACCCACUUUGGCAAUGGUCCCUCGUGCCAAAAAGCACAAAGGGAAAAA_____GCACA____AACGAUAACCACUUCGA ................(((......((((((....)))))).................((((..((((......))))...))))..........................)))...... (-11.93 = -12.60 + 0.67)


| Location | 17,598,983 – 17,599,094 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.30 |
| Shannon entropy | 0.41823 |
| G+C content | 0.39211 |
| Mean single sequence MFE | -29.57 |
| Consensus MFE | -15.93 |
| Energy contribution | -16.30 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.850231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
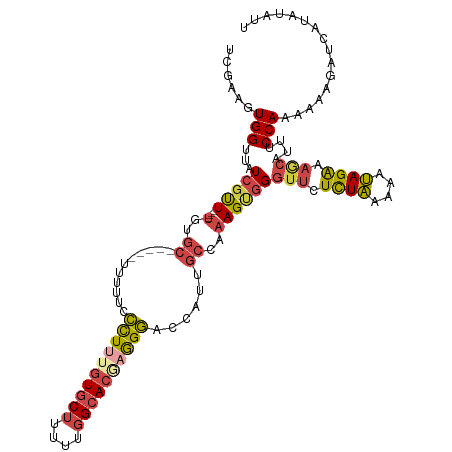
>dm3.chr2R 17598983 111 - 21146708 UCGAAGUGGUUAUCGUU----UGUGC-----UUUUUCCCUUUGUGCUUUUUGGCACGAGGGACCAUUGCCAAAGCGGGUUCUCUAAAAAUAGAAAGCAUUUCCAAAAACGAUUAUAUUUU ..((((((...((((((----(((((-----(((((.((((((((((....))))))))))(((...((....)).)))...........)))))))))......)))))))..)))))) ( -33.30, z-score = -2.93, R) >droAna3.scaffold_13266 10997236 105 - 19884421 CCAUAGUGGUUAUAGCUGGGCUAUAUCAGAUUUUUUCGUUUUGUGCUUUUUUGAAGAUAGCACCAGU-----AGAGGUGUAUUUGAGGACAGGAUG----UCCAACACAGUUAA------ .....(((..((((((...))))))((((((..((((.....(((((.((.....)).)))))....-----.))))...))))))((((.....)----)))..)))......------ ( -25.30, z-score = -1.00, R) >droEre2.scaffold_4845 11747957 110 - 22589142 UCGACGUGGUUAUCGUU----UGUGC------UUUUCCCUUUGUGCUUUUUGGCACGAGGGACCAUUGCCUAAGUGGGUUUUCUAAAAAUAGAAAGCACUACCAAAAAAUAUCAUUUAUU ......((((.......----.((((------((((.((((((((((....)))))))))).(((((.....)))))..............)))))))).))))................ ( -31.70, z-score = -2.90, R) >droYak2.chr2R 9567500 110 + 21139217 UCGGCGUGGUUAUCGUU----UGUGC------UUUUCCCUUUGUGCUUUUUGGCACGUGGGACCAUUGCCAAAGCGGGUUCUCUAAAAAUAGAAAGCACUACCAAAAAAGAUCAUUUAUU ...(.(((((..(((((----(..((------...((((..((((((....)))))).)))).....))..))))))(((.((((....)))).)))))))))................. ( -27.60, z-score = -0.71, R) >droSec1.super_9 924725 111 - 3197100 UCGAAGUGGUUAUCGUU----UGUGC-----UUUUUCCCUUAGUGCUUUUUGGCACGAGGGACCAAUGCCAAAGUGGGUUCUCUAAAAAUAGAAAGCAUUUCCAAAAACGAUCAUAUAUU ........((.((((((----(((((-----((((((((((.(((((....))))))))))).....(((......)))............))))))))......))))))).))..... ( -29.00, z-score = -1.90, R) >droSim1.chr2R 16250334 111 - 19596830 UCGAAGUGGUUAUCGUU----UGUGC-----UUUUUCCCUUAGUGCUUUUUGGCACGAGGGACCAAUGCCAAAGUGGGUUCUCUAGAAAUAGAAAACAUUUCCAAAAACGAUCAUAUAUU (((...(((...(((((----((.((-----....((((((.(((((....))))))))))).....)))))).)))(((.((((....)))).)))....)))....)))......... ( -30.50, z-score = -2.32, R) >consensus UCGAAGUGGUUAUCGUU____UGUGC_____UUUUUCCCUUUGUGCUUUUUGGCACGAGGGACCAUUGCCAAAGUGGGUUCUCUAAAAAUAGAAAGCAUUUCCAAAAAAGAUCAUAUAUU ......(((............................((((((((((....))))))))))................(((.((((....)))).)))....)))................ (-15.93 = -16.30 + 0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:41:29 2011