| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,592,589 – 3,592,690 |
| Length | 101 |
| Max. P | 0.923723 |

| Location | 3,592,589 – 3,592,690 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.71 |
| Shannon entropy | 0.33783 |
| G+C content | 0.39746 |
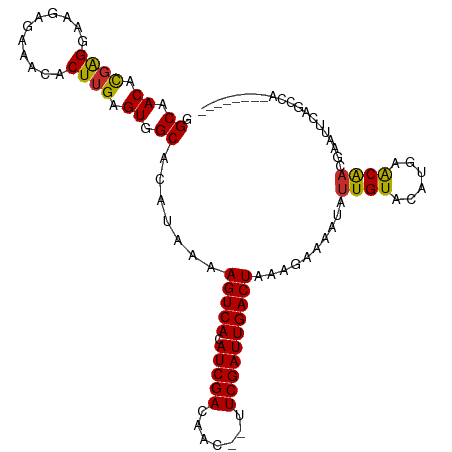
| Mean single sequence MFE | -20.98 |
| Consensus MFE | -15.06 |
| Energy contribution | -14.42 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.923723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3592589 101 - 23011544 GGCAACACGAGGAAGAGAAACACUUGAGUGGCACAUAAAAGUCACAUCGACAAC--UUCGAUUGACUAAAGAAAAUAUUGUACAUGAACAACGAAUUCAGCCA-------- (....).((((...........))))..((((.......(((((.(((((....--.))))))))))...(((....((((......))))....))).))))-------- ( -20.10, z-score = -2.05, R) >droSim1.chr2L 3549622 101 - 22036055 GGCAACACGAGGAAGAGAAACACUUGAGUGGCACAUAAAAGUCACAUCGACAAC--UUCGAUUGACUAAAGAAAAUAUUGUACAUGAACAACGAAUUCAGCCA-------- (....).((((...........))))..((((.......(((((.(((((....--.))))))))))...(((....((((......))))....))).))))-------- ( -20.10, z-score = -2.05, R) >droSec1.super_5 1693457 101 - 5866729 GGCAACACGAGGAAGAGAAACACUUGAGUGGCACAUAAAAGUCACAUCGACAAC--UUCGAUUGACUAAAGAAAAUAUUGUACAUGAACAACGAAUUCAGCCA-------- (....).((((...........))))..((((.......(((((.(((((....--.))))))))))...(((....((((......))))....))).))))-------- ( -20.10, z-score = -2.05, R) >droYak2.chr2L 3584475 101 - 22324452 GGCAACACGAGGAAGAGAAACACUUGAGUGGCACAUAAAAGUCACAUCGACAAC--UUCGAUUGACUAAAGAAAAUAUUGUACAUGAACAACGAAUUCAGCCA-------- (....).((((...........))))..((((.......(((((.(((((....--.))))))))))...(((....((((......))))....))).))))-------- ( -20.10, z-score = -2.05, R) >droEre2.scaffold_4929 3631400 101 - 26641161 GGCAACACGAGGAAGGGAAACACUUGAGUGGCACAUAAAAGUCACAUCGACAAC--UUCGAUUGACUAAAGAAAAUAUUGUACAUGAACAACGAAUUCAGCCA-------- (((....(((((((((....).)))(((((((........))))).)).....)--))))..........(((....((((......))))....))).))).-------- ( -23.10, z-score = -2.91, R) >droAna3.scaffold_12916 5643145 108 - 16180835 AGCAACAUGAGGAAAAGAAACACUUGAGUGGCAUAUAAAAGUCACAUCGACAAC--UUCGAUUGACUAAAGAAAAUAUUGUACAUGAACAAAACCUUACCUUACCAGCAA- .((....(((((........(((....))).........(((((.(((((....--.))))))))))..........((((......)))).......)))))...))..- ( -18.60, z-score = -1.84, R) >dp4.chr4_group3 6540954 108 + 11692001 GGCAACAUGGGGAAGAGAAACACUUGAGUGGCAUAUAAAAGUCACAUCGACAAC--UUCGAUUGACUAAAGAAAAUAUUGUACAU-GGCGAUGGAACACUGAACAUCGUCG (....).(((..((..(((....(((((((((........))))).))))....--)))..))..))).................-(((((((..........))))))). ( -23.10, z-score = -1.53, R) >droPer1.super_1 3640696 108 + 10282868 GGCAACAUGGGGAAGAGAAACACUUGAGUGGCAUAUAAAAGUCACAUCGACAAC--UUCGAUUGACUAAAGAAAAUAUUGUACAU-GGCGAUGGAACACUGAACAUUGUCG (....).(((..((..(((....(((((((((........))))).))))....--)))..))..))).................-(((((((..........))))))). ( -21.00, z-score = -0.95, R) >droWil1.scaffold_180708 2984346 90 + 12563649 GGCAACAUUGGG-GCAGAAACACUUGAGUGGCAUAUAAAAGUCACAUCGACAACUUUUCGAUUGACUAAAGAAAAUAUUGUGUGU--ACGCCG------------------ (((..((((.((-(........))).))))((((((((.(((((.(((((.......))))))))))..........))))))))--..))).------------------ ( -22.60, z-score = -2.08, R) >consensus GGCAACACGAGGAAGAGAAACACUUGAGUGGCACAUAAAAGUCACAUCGACAAC__UUCGAUUGACUAAAGAAAAUAUUGUACAUGAACAACGAAUUCAGCCA________ .((.((.((((...........)))).)).)).......(((((.(((((.......))))))))))..........((((......)))).................... (-15.06 = -14.42 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:14:08 2011