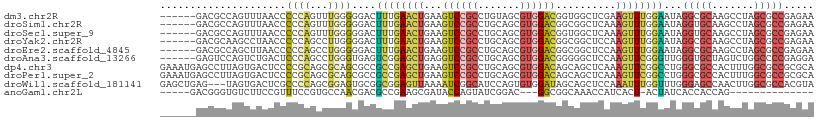
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,510,346 – 17,510,449 |
| Length | 103 |
| Max. P | 0.981059 |

| Location | 17,510,346 – 17,510,449 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 63.55 |
| Shannon entropy | 0.76988 |
| G+C content | 0.60985 |
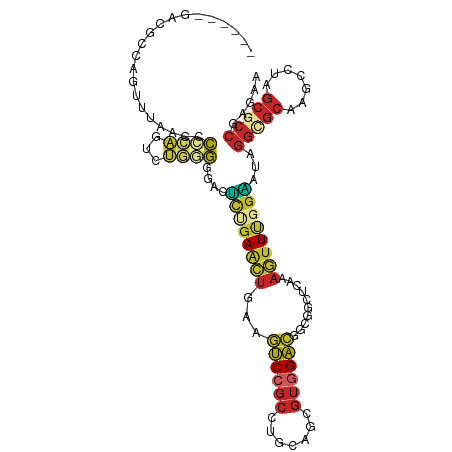
| Mean single sequence MFE | -40.64 |
| Consensus MFE | -16.96 |
| Energy contribution | -16.38 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.75 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.964115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
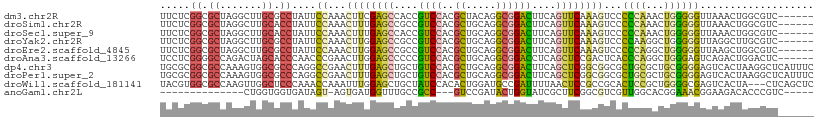
>dm3.chr2R 17510346 103 + 21146708 UUCUCGGCGCUAGGCUUGCGCCUAUUCCAAACUUCGAGCCACCGUCCACGCUACAGGCGGACUUCAGUUCAAAGUCCCCCAAACUGGGGGUUAAACUGGCGUC------ .....((((((((((....)))).......((((.((((....((((..((.....))))))....)))).))))(((((.....))))).......))))))------ ( -34.60, z-score = -1.59, R) >droSim1.chr2R 16158709 103 + 19596830 UUCUCGGCGCUAGGCUUGCACCUAUUCCAAACUUUGAGCCGCCGUCCACGCUGCAGGCGGACUUCAGUUCAAAGUCCCCCAAACUGGGGGUUAAACUGGCGUC------ .....((((((((.................(((((((((.(..((((..((.....))))))..).)))))))))(((((.....))))).....))))))))------ ( -39.30, z-score = -2.41, R) >droSec1.super_9 836381 103 + 3197100 UUCUCGGCGCUAGGCUUGCACCUAUUCCAAACUUUGAGCCACCGUCCACGCUGCAGGCGGACUUCAGUUCAAAGUCCCCCAAACUGGGGGUUAAACUGGCGUC------ .....((((((((.................(((((((((....((((..((.....))))))....)))))))))(((((.....))))).....))))))))------ ( -38.60, z-score = -2.64, R) >droYak2.chr2R 9477623 103 - 21139217 UUCUCGGCGCUAGGCUUGCGCCUAUUCCAAACUUGGAGCCGCCGUCCACGCUGCAGGCGGACUUCAGUUCAAAGUCCCCAAGGCUGGGGGUUAGGCUUGCGUC------ .....(((((.(((((((.((((...(((..(((((.(((((.((....)).)).)))((((((.......)))))))))))..)))))))))))))))))))------ ( -43.20, z-score = -1.52, R) >droEre2.scaffold_4845 11659244 103 + 22589142 UUCUCGGCGCUAGGCUUGCGCCUAUUCCAAACUUGGAGCCGCCGUCCACGCUGCAGGCGGACUUCAGUUCAAAGUCCCCCAGGCUGGGGGUUAAGCUGGCGUC------ .....((((((((.(((((.((((.((((....))))(((((.((....)).)).((.((((((.......)))))).)).))))))).).))))))))))))------ ( -41.60, z-score = -0.86, R) >droAna3.scaffold_13266 8996739 103 + 19884421 UCCUCGGGGCCAGACUAGCACCCAACCCGAACUUGGAGCCCCCGUCCACGCUGCAGGCGGACCUCAGCUCCGACUCACCCAGGCUGGGAGUCAGACUGGACUC------ .....(((((..(.....)..((((.......)))).))))).(((((.((((.(((....)))))))((.(((((.((......))))))).)).)))))..------ ( -35.40, z-score = 0.54, R) >dp4.chr3 4986161 109 + 19779522 UGCGCGGCGCCAAAGUGGCGCCCAGGCCGAACUUUGAGCUGCUGUCCACGCUGCAGGCGGACUUCAGCUCGGCGGCGCUGCGCUGCGGGGAGUCACUAAGGCUCAUUUC .((((((((((((((((((......)))..)))))((((((..((((..((.....))))))..))))))...))))))))))......(((((.....)))))..... ( -54.30, z-score = -1.47, R) >droPer1.super_2 5185302 109 + 9036312 UGCGCGGCGCCAAAGUGGCGCCCAGGCCGAACUUUGAGCUGCUGUCCACGCUGCAGGCGGACUUCAGCUCGGCGGCGCUGCGCUGCGGGGAGUCACUAAGGCUCAUUUC .((((((((((((((((((......)))..)))))((((((..((((..((.....))))))..))))))...))))))))))......(((((.....)))))..... ( -54.30, z-score = -1.47, R) >droWil1.scaffold_181141 1004565 106 - 5303230 UACGUGGCGCCAAGUUGGCUCCCAAACCAAAUUUGGAGCUGCUAUCCACACUGGAUGCCGAUUUUAACUCCGCCGCACUCCGCUGGGGCGAGUCACUA---CUCAGCUC ...((((((...((((((((((.((......)).))))))((.((((.....)))))).......)))).)))))).....((((((...........---)))))).. ( -35.00, z-score = -1.67, R) >anoGam1.chr2L 42465745 86 + 48795086 --------------CUGGUGGUGAUAGU-AGUGAUGGUUUGCCGCC---GUCCGAUACUGGUAUCGCUUCGGCGUCGUUGGCACGGAAACGGAAGACACCCGUC----- --------------..(((((((.((((-(..((((((.....)))---)))...))))).))))))).(((.((.(((..(.((....)))..))))))))..----- ( -30.10, z-score = -0.54, R) >consensus UUCUCGGCGCUAGGCUUGCGCCUAUUCCAAACUUUGAGCCGCCGUCCACGCUGCAGGCGGACUUCAGUUCAAAGUCCCCCAGACUGGGGGUUAAACUGGCGUC______ .....((.((.......)).))....((...((((((((....((((..((.....))))))....))))))))...((((...))))))................... (-16.96 = -16.38 + -0.58)
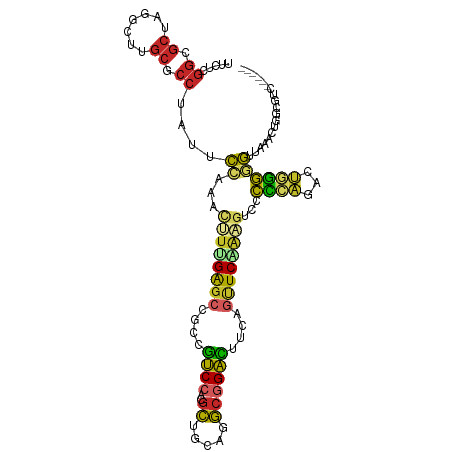
| Location | 17,510,346 – 17,510,449 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 63.55 |
| Shannon entropy | 0.76988 |
| G+C content | 0.60985 |
| Mean single sequence MFE | -42.17 |
| Consensus MFE | -17.74 |
| Energy contribution | -17.59 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.981059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17510346 103 - 21146708 ------GACGCCAGUUUAACCCCCAGUUUGGGGGACUUUGAACUGAAGUCCGCCUGUAGCGUGGACGGUGGCUCGAAGUUUGGAAUAGGCGCAAGCCUAGCGCCGAGAA ------..((((((((((((((((.....)))))...)))))))...(((((((....).))))))))))........(((((..(((((....)))))...))))).. ( -46.30, z-score = -3.65, R) >droSim1.chr2R 16158709 103 - 19596830 ------GACGCCAGUUUAACCCCCAGUUUGGGGGACUUUGAACUGAAGUCCGCCUGCAGCGUGGACGGCGGCUCAAAGUUUGGAAUAGGUGCAAGCCUAGCGCCGAGAA ------....((((.....(((((.....)))))(((((((.(((..(((((((....).))))))..))).)))))))))))....(((((.......)))))..... ( -43.00, z-score = -2.48, R) >droSec1.super_9 836381 103 - 3197100 ------GACGCCAGUUUAACCCCCAGUUUGGGGGACUUUGAACUGAAGUCCGCCUGCAGCGUGGACGGUGGCUCAAAGUUUGGAAUAGGUGCAAGCCUAGCGCCGAGAA ------....((((.....(((((.....)))))(((((((.(((..(((((((....).))))))..))).)))))))))))....(((((.......)))))..... ( -41.00, z-score = -2.14, R) >droYak2.chr2R 9477623 103 + 21139217 ------GACGCAAGCCUAACCCCCAGCCUUGGGGACUUUGAACUGAAGUCCGCCUGCAGCGUGGACGGCGGCUCCAAGUUUGGAAUAGGCGCAAGCCUAGCGCCGAGAA ------.......(((....((((......)))).............(((((((....).)))))))))(((((((....)))).(((((....)))))..)))..... ( -41.30, z-score = -1.53, R) >droEre2.scaffold_4845 11659244 103 - 22589142 ------GACGCCAGCUUAACCCCCAGCCUGGGGGACUUUGAACUGAAGUCCGCCUGCAGCGUGGACGGCGGCUCCAAGUUUGGAAUAGGCGCAAGCCUAGCGCCGAGAA ------...(((.......(((((.....))))).............(((((((....).)))))))))(((((((....)))).(((((....)))))..)))..... ( -45.20, z-score = -2.20, R) >droAna3.scaffold_13266 8996739 103 - 19884421 ------GAGUCCAGUCUGACUCCCAGCCUGGGUGAGUCGGAGCUGAGGUCCGCCUGCAGCGUGGACGGGGGCUCCAAGUUCGGGUUGGGUGCUAGUCUGGCCCCGAGGA ------(.(.((((.(((.(.(((((((((..(.....((((((...(((((((....).))))))...))))))..)..))))))))).).))).)))).).)..... ( -47.90, z-score = -0.59, R) >dp4.chr3 4986161 109 - 19779522 GAAAUGAGCCUUAGUGACUCCCCGCAGCGCAGCGCCGCCGAGCUGAAGUCCGCCUGCAGCGUGGACAGCAGCUCAAAGUUCGGCCUGGGCGCCACUUUGGCGCCGCGCA .............(((......))).((((((.((((..((((((..(((((((....).))))))..))))))......)))))).((((((.....)))))))))). ( -50.10, z-score = -1.38, R) >droPer1.super_2 5185302 109 - 9036312 GAAAUGAGCCUUAGUGACUCCCCGCAGCGCAGCGCCGCCGAGCUGAAGUCCGCCUGCAGCGUGGACAGCAGCUCAAAGUUCGGCCUGGGCGCCACUUUGGCGCCGCGCA .............(((......))).((((((.((((..((((((..(((((((....).))))))..))))))......)))))).((((((.....)))))))))). ( -50.10, z-score = -1.38, R) >droWil1.scaffold_181141 1004565 106 + 5303230 GAGCUGAG---UAGUGACUCGCCCCAGCGGAGUGCGGCGGAGUUAAAAUCGGCAUCCAGUGUGGAUAGCAGCUCCAAAUUUGGUUUGGGAGCCAACUUGGCGCCACGUA ..(((((.---...(((((((((.((......)).))).))))))...))))).......((((........(((((((...))))))).(((.....))).))))... ( -35.10, z-score = 0.06, R) >anoGam1.chr2L 42465745 86 - 48795086 -----GACGGGUGUCUUCCGUUUCCGUGCCAACGACGCCGAAGCGAUACCAGUAUCGGAC---GGCGGCAAACCAUCACU-ACUAUCACCACCAG-------------- -----((((((.....))))))..(((....))).(((((...(((((....)))))..)---)))).............-..............-------------- ( -21.70, z-score = -0.02, R) >consensus ______GACGCCAGUUUAACCCCCAGUCUGGGGGACUCUGAACUGAAGUCCGCCUGCAGCGUGGACGGCGGCUCAAAGUUUGGAAUAGGCGCAAGCCUAGCGCCGAGAA .....................((((...))))....((((((((...((((((.......))))))..........))))))))...(((((.......)))))..... (-17.74 = -17.59 + -0.15)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:41:16 2011