
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,468,769 – 17,468,868 |
| Length | 99 |
| Max. P | 0.856181 |

| Location | 17,468,769 – 17,468,868 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 70.96 |
| Shannon entropy | 0.53244 |
| G+C content | 0.41698 |
| Mean single sequence MFE | -22.12 |
| Consensus MFE | -11.55 |
| Energy contribution | -10.73 |
| Covariance contribution | -0.82 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.856181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17468769 99 - 21146708 --------------------GAGGCUAGCCAAAACUUUUGGCUACCAUCGGCCAUCGUAUCAGCUGAAAGAAAUUUUAAUUAAAUGUCGCCGCAAACGAUGCGGUAAUAUGCAACGAGU-- --------------------((((.(((((((.....))))))))).)).....((((....(((((((....))))).....((((..(((((.....)))))..)))))).))))..-- ( -28.90, z-score = -2.37, R) >droSim1.chr2R 16127907 97 - 19596830 ----------------------GGCUAGCCAAAACUUUUGGCUACCAUCGGCCAUCGUAUCAGCUGAAAGAAAUUUUAAUUAAAUGUCGCCGCAAACGAUGCGGUAAUAUGCAACGAGU-- ----------------------((((((((((.....))))))......)))).((((....(((((((....))))).....((((..(((((.....)))))..)))))).))))..-- ( -28.60, z-score = -2.46, R) >droSec1.super_9 796218 97 - 3197100 ----------------------GGCUAGCCAAAACUUUUGGCUACCAUCGGCCAUCGUAUCAGCUGAAAGAAAUUUUAAUUAAAUGUCGCCGCAAACGAUGCGGUAAUAUGCAACGAGU-- ----------------------((((((((((.....))))))......)))).((((....(((((((....))))).....((((..(((((.....)))))..)))))).))))..-- ( -28.60, z-score = -2.46, R) >droYak2.chr2R 9432231 93 + 21139217 ------------------------CUAGCCAAAACUUUUGGCUACC--CGGCCAUCGUAUCAGCUGAAAGAAAUUUUAAUUAAAUGUCGCCGCAAACGAUGCGGUAAUAUGCAACCAGU-- ------------------------.(((((((.....)))))))..--..............(((((((....))))).....((((..(((((.....)))))..)))))).......-- ( -23.20, z-score = -1.73, R) >droEre2.scaffold_4845 11613999 95 - 22589142 ----------------------GGCUGCCCAAAACUUUUGGCUGCC--CGGCCAUCCUAUCAGCUGAAAGAAAUUUUAAUUAAAUGUCGCCGCAAACGAUGCGGUAAUAUGCAACGAGU-- ----------------------.(.(((......((((..((((..--.((....))...))))..)))).............((((..(((((.....)))))..))))))).)....-- ( -26.50, z-score = -1.51, R) >droAna3.scaffold_13266 5302330 94 - 19884421 --------------------AAAACUAG-GAAAACUUUUCGCC----UCGGCCAUCGUAUCAGCUGAAAGAAAUUUUAAUUAAAUGUCGCCGCAAACGAUGCGGUAAUAUGAAUCGAGU-- --------------------.......(-(((....))))..(----((((...((((((....(((((....)))))..........((((((.....)))))).)))))).))))).-- ( -19.30, z-score = -0.46, R) >dp4.chr3 15751384 86 - 19779522 ----------------------------CCAAAACUUUUGGCC-----CGGCCAUCCUAUCAGCCGAAAGAAAUUUUAAUUAAAUGUCGCCGCAAACGAUGCGGUAAUAUGAAUCGAGC-- ----------------------------......((((((((.-----.((....)).....)))))))).............((((..(((((.....)))))..)))).........-- ( -20.80, z-score = -1.61, R) >droPer1.super_4 3862787 86 + 7162766 ----------------------------CCAAAACUUUUGGCC-----CGGCCAUCCUAUCAGCCGAAAGAAAUUUUAAUUAAAUGUCGCCGCAAACGAUGCGGUAAUAUGAAUCGAGC-- ----------------------------......((((((((.-----.((....)).....)))))))).............((((..(((((.....)))))..)))).........-- ( -20.80, z-score = -1.61, R) >droWil1.scaffold_180745 33328 84 + 2843958 ----------------------------CCAAAACUUUUGGC---------CAAACGUAUCAGCAAAAAGAAAUUUUAAUUAAAUGUCGCCGCAAACGAUGUGGUAAUAUGAAUAUAGAGC ----------------------------......(((((.((---------...........)).)))))...(((((.....((((..(((((.....)))))..)))).....))))). ( -13.40, z-score = -0.62, R) >droVir3.scaffold_12875 2895835 86 + 20611582 ------------------------CGACCGAAAACUAGCUGAUA---UAAACGACGGU-UGCCCAAAAGAAAAUUUUAAUUAAAUGUCGCCGCAAACGAUGCGGUAAUUCGAGC------- ------------------------....((((............---.....(((((.-...))((((.....))))........)))((((((.....))))))..))))...------- ( -15.10, z-score = -0.02, R) >droMoj3.scaffold_6496 15523570 111 + 26866924 AACGGCUUAAAAGCACAAAACCAAAUGUGCGAAACUAGAUAUGC---GCCACGAUGUUGUGCCCAAAAGAAAAUUUUAAUUAAAUGUCGCCGCUAACGAUACGGUAAUUCGAGC------- ..((((......(((((........))))).......(((((((---((.((...)).))))..((((.....))))......)))))))))....(((.........)))...------- ( -20.30, z-score = 0.27, R) >droGri2.scaffold_15245 13970312 77 - 18325388 ------------------------------AAAACUAGAUAUG------CACGACAUU-UGCCUAAGAGAAAAUUUUAAUUAAAUGUCGCCGCAAACGAUGCGGUAAUUCGAGU------- ------------------------------...(((.((....------...((((((-((..(((((.....)))))..))))))))((((((.....))))))...)).)))------- ( -19.90, z-score = -3.10, R) >consensus ________________________CUAGCCAAAACUUUUGGCU_____CGGCCAUCGUAUCAGCUGAAAGAAAUUUUAAUUAAAUGUCGCCGCAAACGAUGCGGUAAUAUGAAACGAGU__ .......................................((((......))))............((((....))))........((..(((((.....)))))..))............. (-11.55 = -10.73 + -0.82)

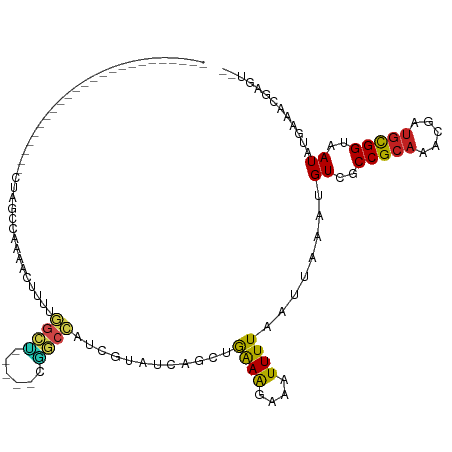
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:41:11 2011