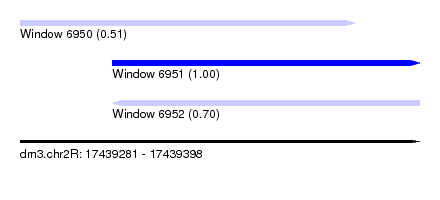
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,439,281 – 17,439,398 |
| Length | 117 |
| Max. P | 0.997816 |

| Location | 17,439,281 – 17,439,379 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 75.86 |
| Shannon entropy | 0.43923 |
| G+C content | 0.58714 |
| Mean single sequence MFE | -18.99 |
| Consensus MFE | -13.20 |
| Energy contribution | -13.04 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.512049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17439281 98 + 21146708 --CCGUUCCCUCAGCCGCGCCCCA-CGGAAACCAUCCCACCACCCGCUGCAGUUGCGCUUAUUUGCCUUCGCCUUCGUGACGCCACGAUGCGAGCUUAGAA --.......((.(((.((((..((-(((...............))).)).....))))..........((((..(((((....))))).))))))).)).. ( -23.06, z-score = -0.71, R) >droSim1.chr2R 16098514 98 + 19596830 --ACGUUCCCCCAGCCCUGCCCCU-CGCAAACCAUCCCACCACCCGCUGCAGUUGCGCUUAUUUGCCUUCGCCUUCGUGACGCCACGAUGCGAGCUUAGAA --.(((..(..((((..(((....-.)))................))))..)..)))....((((..(((((..(((((....))))).)))))..)))). ( -19.37, z-score = -0.92, R) >droSec1.super_9 767058 98 + 3197100 --ACGUUCCCCCAGCCCUGCCCCU-CGCAAGCCAUCCCACCACCCGCUGCAGUUGCGCUUAUUUGCCUUCGCCUUCGUGACGCCACGAUGCGAGCUUAGAA --.(((..(..((((..(((....-.))).(......).......))))..)..)))....((((..(((((..(((((....))))).)))))..)))). ( -20.30, z-score = -0.67, R) >droYak2.chr2R 9402035 101 - 21139217 CCCCCUUUUCCAAGCCCUGCCCCCACUGAAACCAUCCCACCACCCGCUGCAGUUGCGCUUAUUUGCCUUCGCCUUCGUGACGCCACGAUGCGAGCAUAGAA ...........(((((((((......((............))......))))..).))))...(((..((((..(((((....))))).)))))))..... ( -18.30, z-score = -1.36, R) >droEre2.scaffold_4845 11584394 86 + 22589142 --------------CCCUGCCCCU-CUGAAACCAACCCACCACCCGCUGCAGUUGCGCUUAUUUGCCUUCGCCUUCGUGACGCCACGAUGCGAGCAUAGAA --------------.........(-(((.................((.((....)))).....(((..((((..(((((....))))).))))))))))). ( -18.60, z-score = -1.76, R) >dp4.chr3 15720185 76 + 19779522 -------------------------UACCAUCCGCCCUCUCAGCAGUUGCAGUUGCGAUUAUUUGCCUUCGUCUUCGUGACGCCACGACUCGAGCAUAGAA -------------------------............(((..((((((((....))))))........(((...(((((....)))))..)))))..))). ( -16.10, z-score = -0.32, R) >droPer1.super_4 3831242 76 - 7162766 -------------------------UACCAUCCGCCCUCUCAGCAGUUGCAGUUGCGAUUAUUUGCCUUCGUCUUCGUGACGCCACGACGCGAGCAUAGAA -------------------------............(((..((((((((....))))))........((((..(((((....))))).))))))..))). ( -17.20, z-score = -0.23, R) >consensus ___C_UU__C__AGCCCUGCCCCU_CGCAAACCAUCCCACCACCCGCUGCAGUUGCGCUUAUUUGCCUUCGCCUUCGUGACGCCACGAUGCGAGCAUAGAA .............................................((.((((..........))))..((((..(((((....))))).))))))...... (-13.20 = -13.04 + -0.16)
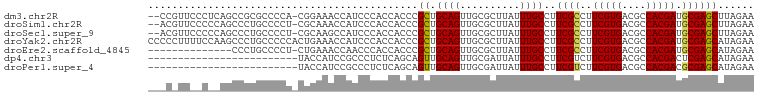
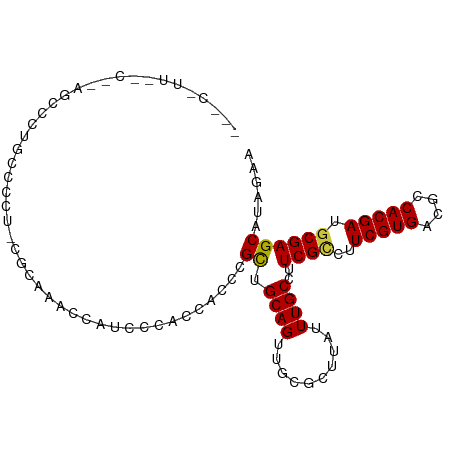
| Location | 17,439,308 – 17,439,398 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Shannon entropy | 0.03485 |
| G+C content | 0.61111 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -34.46 |
| Energy contribution | -34.66 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.997816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17439308 90 + 21146708 ACCAUCCCACCACCCGCUGCAGUUGCGCUUAUUUGCCUUCGCCUUCGUGACGCCACGAUGCGAGCUUAGAACAGCGCGGCAAUAGCGGCC .............((((((..((((((((..((((..(((((..(((((....))))).)))))..))))..))))))))..)))))).. ( -35.80, z-score = -3.71, R) >droSim1.chr2R 16098541 90 + 19596830 ACCAUCCCACCACCCGCUGCAGUUGCGCUUAUUUGCCUUCGCCUUCGUGACGCCACGAUGCGAGCUUAGAACAGCGCGGCAAUAGCGGCC .............((((((..((((((((..((((..(((((..(((((....))))).)))))..))))..))))))))..)))))).. ( -35.80, z-score = -3.71, R) >droSec1.super_9 767085 90 + 3197100 GCCAUCCCACCACCCGCUGCAGUUGCGCUUAUUUGCCUUCGCCUUCGUGACGCCACGAUGCGAGCUUAGAACAGCGCGGCAAUAGCGGCC .............((((((..((((((((..((((..(((((..(((((....))))).)))))..))))..))))))))..)))))).. ( -35.80, z-score = -3.26, R) >droYak2.chr2R 9402065 90 - 21139217 ACCAUCCCACCACCCGCUGCAGUUGCGCUUAUUUGCCUUCGCCUUCGUGACGCCACGAUGCGAGCAUAGAACAGCGCGGCAAUAGCGGCC .............((((((..((((((((....(((..((((..(((((....))))).)))))))......))))))))..)))))).. ( -35.90, z-score = -3.73, R) >droEre2.scaffold_4845 11584409 90 + 22589142 ACCAACCCACCACCCGCUGCAGUUGCGCUUAUUUGCCUUCGCCUUCGUGACGCCACGAUGCGAGCAUAGAACAGCGCGGCAAUAGCAGCC ...............(((((.((((((((....(((..((((..(((((....))))).)))))))......))))))))....))))). ( -33.60, z-score = -3.82, R) >consensus ACCAUCCCACCACCCGCUGCAGUUGCGCUUAUUUGCCUUCGCCUUCGUGACGCCACGAUGCGAGCUUAGAACAGCGCGGCAAUAGCGGCC .............((((((..((((((((..((((..(((((..(((((....))))).)))))..))))..))))))))..)))))).. (-34.46 = -34.66 + 0.20)


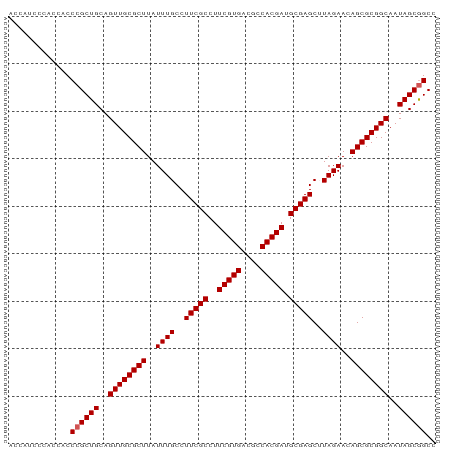
| Location | 17,439,308 – 17,439,398 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Shannon entropy | 0.03485 |
| G+C content | 0.61111 |
| Mean single sequence MFE | -37.38 |
| Consensus MFE | -36.94 |
| Energy contribution | -36.62 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.699344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
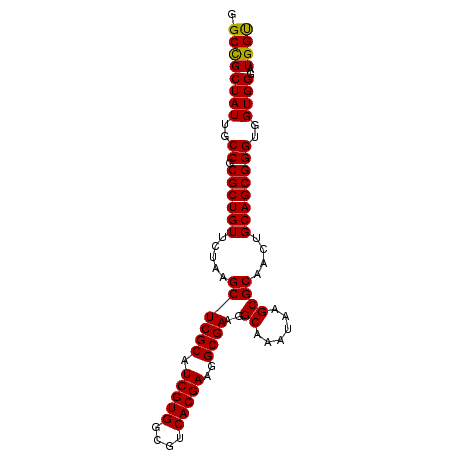
>dm3.chr2R 17439308 90 - 21146708 GGCCGCUAUUGCCGCGCUGUUCUAAGCUCGCAUCGUGGCGUCACGAAGGCGAAGGCAAAUAAGCGCAACUGCAGCGGGUGGUGGGAUGGU .((((((((..((.((((((.....((((((.(((((....)))))..))))..((......))))....))))))))..))))..)))) ( -37.00, z-score = -1.02, R) >droSim1.chr2R 16098541 90 - 19596830 GGCCGCUAUUGCCGCGCUGUUCUAAGCUCGCAUCGUGGCGUCACGAAGGCGAAGGCAAAUAAGCGCAACUGCAGCGGGUGGUGGGAUGGU .((((((((..((.((((((.....((((((.(((((....)))))..))))..((......))))....))))))))..))))..)))) ( -37.00, z-score = -1.02, R) >droSec1.super_9 767085 90 - 3197100 GGCCGCUAUUGCCGCGCUGUUCUAAGCUCGCAUCGUGGCGUCACGAAGGCGAAGGCAAAUAAGCGCAACUGCAGCGGGUGGUGGGAUGGC .((((((((..((.((((((.....((((((.(((((....)))))..))))..((......))))....))))))))..))))..)))) ( -38.00, z-score = -1.19, R) >droYak2.chr2R 9402065 90 + 21139217 GGCCGCUAUUGCCGCGCUGUUCUAUGCUCGCAUCGUGGCGUCACGAAGGCGAAGGCAAAUAAGCGCAACUGCAGCGGGUGGUGGGAUGGU .((((((((..((.((((((....(((((((.(((((....)))))..))))..((......)))))...))))))))..))))..)))) ( -38.60, z-score = -1.38, R) >droEre2.scaffold_4845 11584409 90 - 22589142 GGCUGCUAUUGCCGCGCUGUUCUAUGCUCGCAUCGUGGCGUCACGAAGGCGAAGGCAAAUAAGCGCAACUGCAGCGGGUGGUGGGUUGGU .....((((..((.((((((....(((((((.(((((....)))))..))))..((......)))))...))))))))..))))...... ( -36.30, z-score = -0.85, R) >consensus GGCCGCUAUUGCCGCGCUGUUCUAAGCUCGCAUCGUGGCGUCACGAAGGCGAAGGCAAAUAAGCGCAACUGCAGCGGGUGGUGGGAUGGU .((((((((..((.((((((.....((((((.(((((....)))))..))))..((......))))....))))))))..))))..)))) (-36.94 = -36.62 + -0.32)

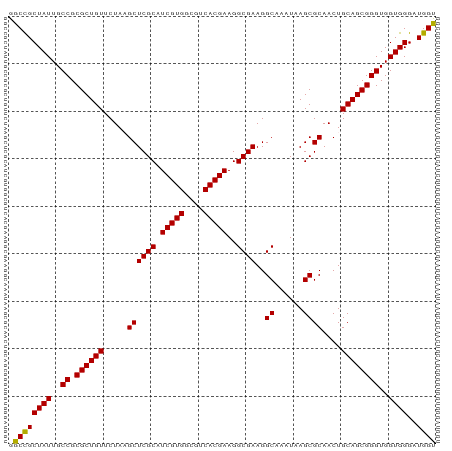
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:41:10 2011