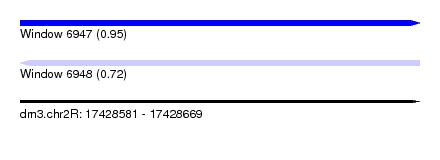
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,428,581 – 17,428,669 |
| Length | 88 |
| Max. P | 0.954557 |

| Location | 17,428,581 – 17,428,669 |
|---|---|
| Length | 88 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 62.57 |
| Shannon entropy | 0.68256 |
| G+C content | 0.48742 |
| Mean single sequence MFE | -25.64 |
| Consensus MFE | -9.16 |
| Energy contribution | -11.61 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.954557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
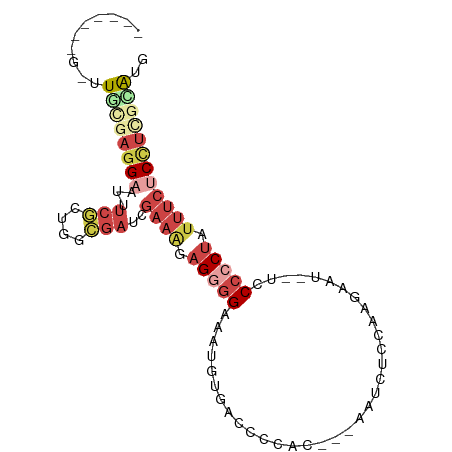
>dm3.chr2R 17428581 88 + 21146708 -------GGUUGCGAGGAAUUUCGCUGGAGAUCGAAAGAGGGGAAUUGUGGUCCCACCACAAUCUCCAAGAAU--UCCCACUAUUUCUCUUCGUAUG -------...((((((((.......(((.((((....))(((((.((((((.....)))))))))))......--))))).......)))))))).. ( -30.44, z-score = -2.49, R) >droPer1.super_4 3820552 95 - 7162766 AGAAUCGGGGUAUUCUGAAGGUCAGAGAUGCUUCCAACUGCAGCCUCCUUUUUCCUUCC--AUCAUCGAACAUCGUGCAUCUAAAAUUCUCCGUGCG .(...(((((.(((..(((((...(((.(((........)))..)))......))))).--..((.((.....)))).......))).)))))..). ( -18.10, z-score = 0.51, R) >droEre2.scaffold_4845 11573660 85 + 22589142 ------AGCUUGC-AGGAAUUUCGCUGGCGAUCGAAGGAGGGGAAAUGUAGCCCCGC---AAUCUCCAAGAAU--UGCCCCUAUUUCCCCUCCGAUG ------.(((.((-.........)).))).......(((((((((..((((....((---((((.....).))--)))..))))))))))))).... ( -27.90, z-score = -1.33, R) >droYak2.chr2R 9390846 72 - 21139217 -------AAUUGAGAGGAAUUUCGCUAGCGAUCGAAAUAGGGGAA----------------AUCACCAAGAAU--UCCCCCUAUUUCCCCUUCAACG -------..(((((.((....(((....)))..((((((((((((----------------.((.....)).)--).)))))))))))).))))).. ( -23.00, z-score = -3.44, R) >droSec1.super_9 756453 83 + 3197100 ---------UUGCGAGGAAUUUCGCUGGCAAUCGAAAGAGGGGAAAUGUGACCCCAU---AAUUUCCAAGAAU--UCCCCCUAUUUCUCCUCGCGUG ---------..(((((((.............((....))((((((.((((....)))---)..(((...))))--))))).......)))))))... ( -25.20, z-score = -2.05, R) >droSim1.chr2R 16087849 83 + 19596830 ---------UUGCGAGGAAUUUCGCUGGCGAUCGAAAGAGGGGAAAUGUGACCCCAC---AAUUUCCAAGAAU--UCCCCCUAUUUCUCCUCGCGUG ---------..(((((((...(((....)))..((((..((((((.((((....)))---)..(((...))))--)))))...)))))))))))... ( -29.20, z-score = -2.92, R) >consensus _______G_UUGCGAGGAAUUUCGCUGGCGAUCGAAAGAGGGGAAAUGUGACCCCAC___AAUCUCCAAGAAU__UCCCCCUAUUUCUCCUCGCAUG ..........((((((((...(((....)))..((((.(((((..................................))))).)))))))))))).. ( -9.16 = -11.61 + 2.45)

| Location | 17,428,581 – 17,428,669 |
|---|---|
| Length | 88 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 62.57 |
| Shannon entropy | 0.68256 |
| G+C content | 0.48742 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -7.70 |
| Energy contribution | -9.53 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.718802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17428581 88 - 21146708 CAUACGAAGAGAAAUAGUGGGA--AUUCUUGGAGAUUGUGGUGGGACCACAAUUCCCCUCUUUCGAUCUCCAGCGAAAUUCCUCGCAACC------- ................((((((--((((((((((((((.((.((((.......)))).))...)))))))))).).)))))).)))....------- ( -29.90, z-score = -2.70, R) >droPer1.super_4 3820552 95 + 7162766 CGCACGGAGAAUUUUAGAUGCACGAUGUUCGAUGAU--GGAAGGAAAAAGGAGGCUGCAGUUGGAAGCAUCUCUGACCUUCAGAAUACCCCGAUUCU ....(((.(.(((((.....((((.....)).))..--.(((((.....(((((.(((........))))))))..)))))))))).).)))..... ( -20.60, z-score = 0.70, R) >droEre2.scaffold_4845 11573660 85 - 22589142 CAUCGGAGGGGAAAUAGGGGCA--AUUCUUGGAGAUU---GCGGGGCUACAUUUCCCCUCCUUCGAUCGCCAGCGAAAUUCCU-GCAAGCU------ ....(((((((((((..(((((--(((......))))---))....))..))))))))))).......((..(((.......)-))..)).------ ( -28.00, z-score = -0.30, R) >droYak2.chr2R 9390846 72 + 21139217 CGUUGAAGGGGAAAUAGGGGGA--AUUCUUGGUGAU----------------UUCCCCUAUUUCGAUCGCUAGCGAAAUUCCUCUCAAUU------- .(((((..((((((((((((((--(((......)))----------------))))))))))))..(((....)))....))..))))).------- ( -30.80, z-score = -4.17, R) >droSec1.super_9 756453 83 - 3197100 CACGCGAGGAGAAAUAGGGGGA--AUUCUUGGAAAUU---AUGGGGUCACAUUUCCCCUCUUUCGAUUGCCAGCGAAAUUCCUCGCAA--------- ...(((((((((((.(((((((--(((((((......---.)))))....))))))))).))))..(((....)))...)))))))..--------- ( -29.40, z-score = -2.39, R) >droSim1.chr2R 16087849 83 - 19596830 CACGCGAGGAGAAAUAGGGGGA--AUUCUUGGAAAUU---GUGGGGUCACAUUUCCCCUCUUUCGAUCGCCAGCGAAAUUCCUCGCAA--------- ...(((((((((((.(((((((--((((...)))..(---(((....)))))))))))).))))..(((....)))...)))))))..--------- ( -32.50, z-score = -2.89, R) >consensus CACACGAGGAGAAAUAGGGGGA__AUUCUUGGAGAUU___GUGGGGCCACAUUUCCCCUCUUUCGAUCGCCAGCGAAAUUCCUCGCAA_C_______ ......((((((((.(((((((....((((............))))......))))))).))))..(((....)))...)))).............. ( -7.70 = -9.53 + 1.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:41:07 2011