| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,367,444 – 17,367,552 |
| Length | 108 |
| Max. P | 0.984631 |

| Location | 17,367,444 – 17,367,552 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 60.45 |
| Shannon entropy | 0.70793 |
| G+C content | 0.36640 |
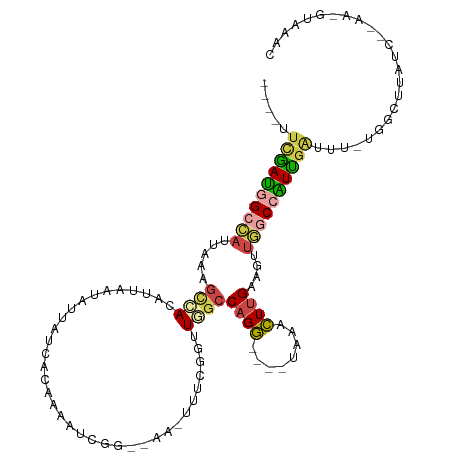
| Mean single sequence MFE | -23.69 |
| Consensus MFE | -8.93 |
| Energy contribution | -9.35 |
| Covariance contribution | 0.42 |
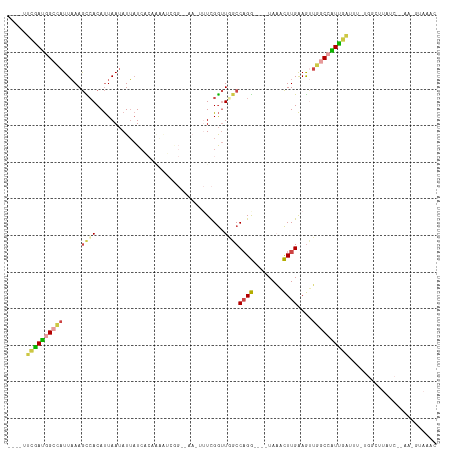
| Combinations/Pair | 1.56 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.984631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
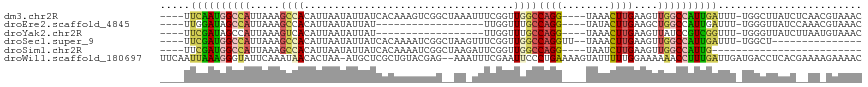
>dm3.chr2R 17367444 108 + 21146708 ----UUCAAUGGCCAUUAAAGCCACAUUAAUAUUAUCACAAAGUCGGCUAAAUUUCGGUUGGCCAGG----UAAACUUGAAGUUGGCCAUUGAUUU-UGGCUUAUCUCAACGUAAAC ----.(((((((((.....((((((.................)).)))).((((((((....))(((----....))))))))))))))))))...-.................... ( -25.43, z-score = -1.66, R) >droEre2.scaffold_4845 11512112 90 + 22589142 ----UUGGAUAGCCAUUAAAGCCACAUUAAUAUUAU------------------UUGGUUUGCCAGG----UAUACUUGAAGCUGGCCAUUGAUUU-UGGGUUAUCCAAACGUAAAC ----(((((((((((((((.(((((.((((...(((------------------((((....)))))----))...)))).).))))..)))))..-..))))))))))........ ( -27.50, z-score = -3.11, R) >droYak2.chr2R 9326349 90 - 21139217 ----UUCGAUAGCCAUUAAAGUCACAUUAAUAUUAU------------------UUGGUUUGCCAGG----UAAACUUGAAGUUAUCCGUCGGUUU-UGGGUUAUCUUAAUGUAAAC ----...(((((((..((((..(((........(((------------------((((....)))))----))(((.....)))....)).)..))-)))))))))........... ( -18.20, z-score = -1.35, R) >droSec1.super_9 697378 95 + 3197100 ----UUCGAUGGCCAUUAAAGCCACAUUAAUAUUAUCACAAAAUCGGCUAAGUUUCGGUUGGCCAGGUU--UAAACUUGAAGUUGGCCAUUGAUUU-UGGCU--------------- ----...(.((((.......)))))....................(((((((..(((((.((((((.((--((....)))).))))))))))).))-)))))--------------- ( -26.50, z-score = -2.12, R) >droSim1.chr2R 16026233 84 + 19596830 ----UUCGAUGGCCAUUAAAGCCACAUUAAUAUUAUCACAAAAUCGGCUAAGAUUCGGUUGGCCAGG----UAAUCUUGAAGUUGGCCAUUG------------------------- ----..(((((((((.....((((.............((..((((......))))..))))))((((----....))))....)))))))))------------------------- ( -22.31, z-score = -2.02, R) >droWil1.scaffold_180697 1684551 114 + 4168966 UUCAAUUAAAGGGUAUUCAAAUAACACUAA-AUGCUCGCUGUACGAG--AAAUUUCGAAUUCCCUGAAAAGUAUUUUUGGAAAAAACCUUUGAUUGAUGACCUCACGAAAAGAAAAC .(((((((((((..................-...((((.....))))--...(((((((....((....))....)))))))....))))))))))).................... ( -22.20, z-score = -1.95, R) >consensus ____UUCGAUGGCCAUUAAAGCCACAUUAAUAUUAUCACAAAAUCGG__AA_UUUCGGUUGGCCAGG____UAAACUUGAAGUUGGCCAUUGAUUU_UGGCUUAUC__AA_GUAAAC .....((((((((((.....((((...................................))))((((........))))....))))))))))........................ ( -8.93 = -9.35 + 0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:58 2011