| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,343,510 – 17,343,617 |
| Length | 107 |
| Max. P | 0.969884 |

| Location | 17,343,510 – 17,343,617 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 89.28 |
| Shannon entropy | 0.20088 |
| G+C content | 0.48561 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -22.95 |
| Energy contribution | -23.03 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
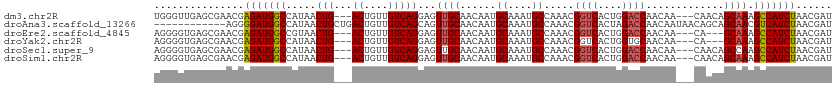
>dm3.chr2R 17343510 107 + 21146708 UGGGUUGAGCGAACGAGAUGGCCAUAACUG---ACUGUUGUCAGGAGUUGCAACAAUGCAAAUGCCAAACGGUCACUGGACCAACAA---CAACAGCAAAGCCAUCUAACGAU .........((....(((((((......((---.(((((((..((..(((((....)))))...))....((((....))))....)---))))))))..)))))))..)).. ( -32.40, z-score = -2.32, R) >droAna3.scaffold_13266 5159859 101 + 19884421 ------------AGGGGAUGGCCAUAACUGCUGACUGUUGUCAGCAGUUGCAACAAUGCAAAUGCCAAACGGUCACUAGACCAACAAUAACAGCAGCAACGUCAUCUAACGAU ------------...(((((((......(((((.(((((((..(((.(((((....))))).))).....((((....))))....))))))))))))..)))))))...... ( -35.40, z-score = -3.92, R) >droEre2.scaffold_4845 11488350 104 + 22589142 AGGGGUGAGCGAACGAGAUGGCCGUAACUG---ACUGUUGUCAGGAGUUGCAACAAUGCAAAUGCCAAACGGUCACUGGACCAACAA---CA---GCAAAGCCAUCUAACGAU .........((....(((((((......((---.(((((((..((..(((((....)))))...))....((((....)))).))))---))---)))..)))))))..)).. ( -32.40, z-score = -1.82, R) >droYak2.chr2R 9298072 104 - 21139217 AGGGGUGAGCGAACGAGAUGGCCAUAACUG---ACUGUUGUCAGGAGUUGCAACAAUGCAAAUGCCAAACGGUCACUGGUGCAACAA---CA---GCAAAGCCAUCUAACGAU ..(((((.((..((.((.(((((.....((---((....))))((..(((((....)))))...))....))))))).))((.....---..---))...)))))))...... ( -30.70, z-score = -1.14, R) >droSec1.super_9 673835 107 + 3197100 AGGGGUGAGCGAACGAGAUGGCCAUAACUG---ACUGUUGUCAGGAGUUGCAACAAUGCAAAUGCCAAACGGUCACUGGACCAACAA---CAACAGCCAAGCCAUCUAACGAU .........((....(((((((......((---.(((((((..((..(((((....)))))...))....((((....))))....)---)))))).)).)))))))..)).. ( -32.00, z-score = -1.79, R) >droSim1.chr2R 16000663 107 + 19596830 AGGGGUGAGCGAACGAGAUGGCCAUAACUG---ACUGUUGUCAGGAGUUGCAACAAUGCAAAUGCCAAACGGUCACUGGACCAACAA---CAACAGCAAAGCCAUCUAACGAU .........((....(((((((......((---.(((((((..((..(((((....)))))...))....((((....))))....)---))))))))..)))))))..)).. ( -32.40, z-score = -2.16, R) >consensus AGGGGUGAGCGAACGAGAUGGCCAUAACUG___ACUGUUGUCAGGAGUUGCAACAAUGCAAAUGCCAAACGGUCACUGGACCAACAA___CAACAGCAAAGCCAUCUAACGAU ...............(((((((.(((((........)))))..((..(((((....)))))...))....((((....))))..................)))))))...... (-22.95 = -23.03 + 0.08)
| Location | 17,343,510 – 17,343,617 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 89.28 |
| Shannon entropy | 0.20088 |
| G+C content | 0.48561 |
| Mean single sequence MFE | -35.90 |
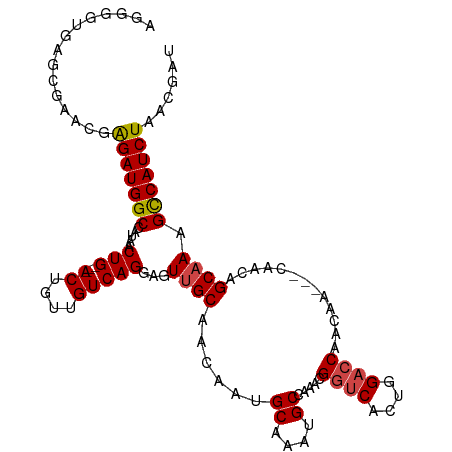
| Consensus MFE | -24.97 |
| Energy contribution | -26.47 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.969884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17343510 107 - 21146708 AUCGUUAGAUGGCUUUGCUGUUG---UUGUUGGUCCAGUGACCGUUUGGCAUUUGCAUUGUUGCAACUCCUGACAACAGU---CAGUUAUGGCCAUCUCGUUCGCUCAACCCA ..((..((((((((..(((((((---(((((((((....))))....((...(((((....)))))..)).)))))))).---))))...))))))))....))......... ( -37.00, z-score = -3.18, R) >droAna3.scaffold_13266 5159859 101 - 19884421 AUCGUUAGAUGACGUUGCUGCUGUUAUUGUUGGUCUAGUGACCGUUUGGCAUUUGCAUUGUUGCAACUGCUGACAACAGUCAGCAGUUAUGGCCAUCCCCU------------ .......((((.(((.(((((((..((((((((((....))))(.(..(((.(((((....))))).)))..))))))))))))))).)))..))))....------------ ( -38.90, z-score = -3.90, R) >droEre2.scaffold_4845 11488350 104 - 22589142 AUCGUUAGAUGGCUUUGC---UG---UUGUUGGUCCAGUGACCGUUUGGCAUUUGCAUUGUUGCAACUCCUGACAACAGU---CAGUUACGGCCAUCUCGUUCGCUCACCCCU ..((..((((((((..((---((---(((((((((....))))(((.((...(((((....)))))..)).)))))))).---))))...))))))))....))......... ( -34.00, z-score = -2.57, R) >droYak2.chr2R 9298072 104 + 21139217 AUCGUUAGAUGGCUUUGC---UG---UUGUUGCACCAGUGACCGUUUGGCAUUUGCAUUGUUGCAACUCCUGACAACAGU---CAGUUAUGGCCAUCUCGUUCGCUCACCCCU ..((..((((((((..((---((---((((((((((((.......))))...(((((....)))))....)).)))))).---))))...))))))))....))......... ( -30.90, z-score = -1.72, R) >droSec1.super_9 673835 107 - 3197100 AUCGUUAGAUGGCUUGGCUGUUG---UUGUUGGUCCAGUGACCGUUUGGCAUUUGCAUUGUUGCAACUCCUGACAACAGU---CAGUUAUGGCCAUCUCGUUCGCUCACCCCU ..((..(((((((((((((((((---(((((((((....))))....((...(((((....)))))..)).)))))))).---)))))).))))))))....))......... ( -37.60, z-score = -2.98, R) >droSim1.chr2R 16000663 107 - 19596830 AUCGUUAGAUGGCUUUGCUGUUG---UUGUUGGUCCAGUGACCGUUUGGCAUUUGCAUUGUUGCAACUCCUGACAACAGU---CAGUUAUGGCCAUCUCGUUCGCUCACCCCU ..((..((((((((..(((((((---(((((((((....))))....((...(((((....)))))..)).)))))))).---))))...))))))))....))......... ( -37.00, z-score = -3.33, R) >consensus AUCGUUAGAUGGCUUUGCUGUUG___UUGUUGGUCCAGUGACCGUUUGGCAUUUGCAUUGUUGCAACUCCUGACAACAGU___CAGUUAUGGCCAUCUCGUUCGCUCACCCCU .......(((((((..((((......(((((((((....))))(((.((...(((((....)))))..)).))))))))....))))...)))))))................ (-24.97 = -26.47 + 1.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:50 2011