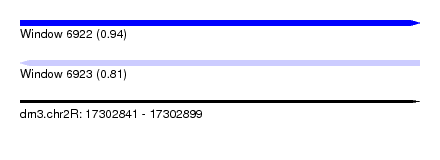
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,302,841 – 17,302,899 |
| Length | 58 |
| Max. P | 0.940549 |

| Location | 17,302,841 – 17,302,899 |
|---|---|
| Length | 58 |
| Sequences | 5 |
| Columns | 59 |
| Reading direction | forward |
| Mean pairwise identity | 77.40 |
| Shannon entropy | 0.39267 |
| G+C content | 0.45507 |
| Mean single sequence MFE | -13.76 |
| Consensus MFE | -11.60 |
| Energy contribution | -11.52 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.940549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
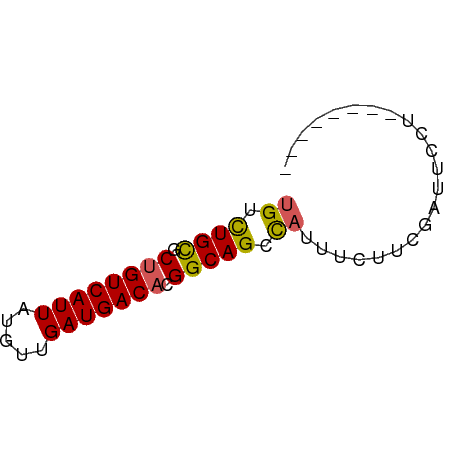
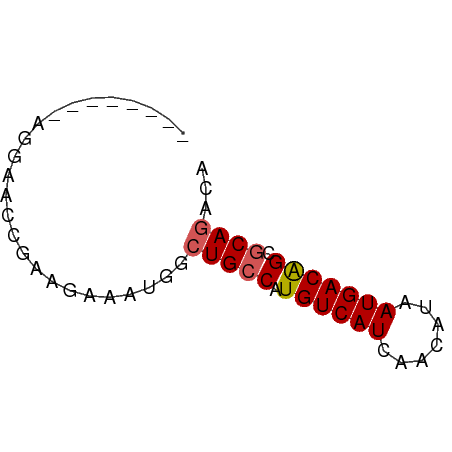
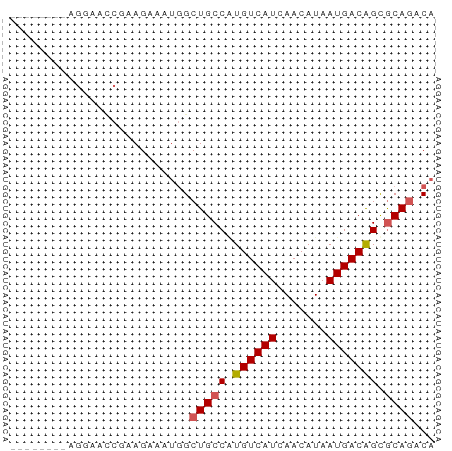
>dm3.chr2R 17302841 58 + 21146708 UGUCUGCACUGUCAUUAUGUUGAUGACAUGGCAGCCAUUUCUU-GUUUCCUCGAUUCCA ((.((((..(((((((.....)))))))..)))).))......-............... ( -13.80, z-score = -1.61, R) >droSec1.super_9 633808 50 + 3197100 UGUCUGCGCUGUCAUUAUGUUGAUGACAUGGCAGCCAUUUCUU-GGCUCCU-------- .........(((((((.....))))))).((.(((((.....)-)))))).-------- ( -15.80, z-score = -1.98, R) >droYak2.chr2R 9255292 51 - 21139217 UGUCUGCGCUGUCAUUAUGUUGAUGACACGGCAGCCAUUUCUUCGAUUCCU-------- ((.((((..(((((((.....)))))))..)))).))..............-------- ( -13.40, z-score = -1.66, R) >droEre2.scaffold_4845 11447332 51 + 22589142 UGUCUGCGCUGUCAUUAUGUUGAUGACACGGCAGCCAUUUCUUCGGUUCCU-------- ((.((((..(((((((.....)))))))..)))).))..............-------- ( -13.40, z-score = -1.27, R) >dp4.chr3 15575857 57 + 19779522 UGUUUGUGCCGUCAUUAUGUUGAUGACA-GGCAGCUUUUACUUCUAGAUUGGAAUCCA- ......((((((((((.....)))))).-))))........((((.....))))....- ( -12.40, z-score = -0.78, R) >consensus UGUCUGCGCUGUCAUUAUGUUGAUGACACGGCAGCCAUUUCUUCGAUUCCU________ ((.((((..(((((((.....)))))))..)))).))...................... (-11.60 = -11.52 + -0.08)

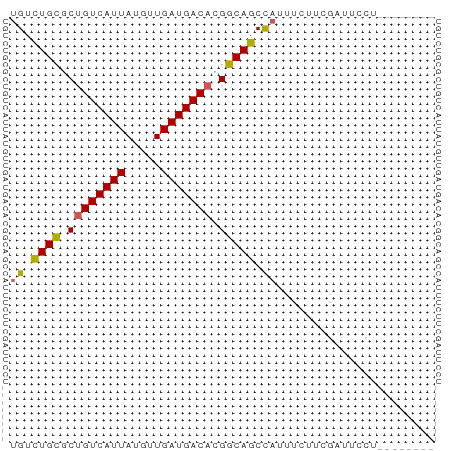
| Location | 17,302,841 – 17,302,899 |
|---|---|
| Length | 58 |
| Sequences | 5 |
| Columns | 59 |
| Reading direction | reverse |
| Mean pairwise identity | 77.40 |
| Shannon entropy | 0.39267 |
| G+C content | 0.45507 |
| Mean single sequence MFE | -12.40 |
| Consensus MFE | -8.98 |
| Energy contribution | -9.22 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.806000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
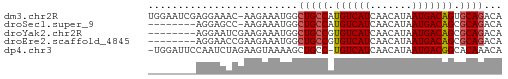
>dm3.chr2R 17302841 58 - 21146708 UGGAAUCGAGGAAAC-AAGAAAUGGCUGCCAUGUCAUCAACAUAAUGACAGUGCAGACA .....((..(....)-..))..((.((((..((((((.......))))))..)))).)) ( -14.00, z-score = -1.75, R) >droSec1.super_9 633808 50 - 3197100 --------AGGAGCC-AAGAAAUGGCUGCCAUGUCAUCAACAUAAUGACAGCGCAGACA --------.((((((-(.....))))).)).((((((.......))))))......... ( -14.20, z-score = -2.03, R) >droYak2.chr2R 9255292 51 + 21139217 --------AGGAAUCGAAGAAAUGGCUGCCGUGUCAUCAACAUAAUGACAGCGCAGACA --------..............((.((((..((((((.......))))))..)))).)) ( -11.20, z-score = -0.75, R) >droEre2.scaffold_4845 11447332 51 - 22589142 --------AGGAACCGAAGAAAUGGCUGCCGUGUCAUCAACAUAAUGACAGCGCAGACA --------..............((.((((..((((((.......))))))..)))).)) ( -11.20, z-score = -0.81, R) >dp4.chr3 15575857 57 - 19779522 -UGGAUUCCAAUCUAGAAGUAAAAGCUGCC-UGUCAUCAACAUAAUGACGGCACAAACA -(((((....))))).........(.((((-.(((((.......))))))))))..... ( -11.40, z-score = -1.55, R) >consensus ________AGGAACCGAAGAAAUGGCUGCCAUGUCAUCAACAUAAUGACAGCGCAGACA .........................((((..((((((.......))))))..))))... ( -8.98 = -9.22 + 0.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:46 2011