
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,302,430 – 17,302,521 |
| Length | 91 |
| Max. P | 0.552305 |

| Location | 17,302,430 – 17,302,520 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 80.83 |
| Shannon entropy | 0.35997 |
| G+C content | 0.44811 |
| Mean single sequence MFE | -16.33 |
| Consensus MFE | -10.61 |
| Energy contribution | -11.92 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.552305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
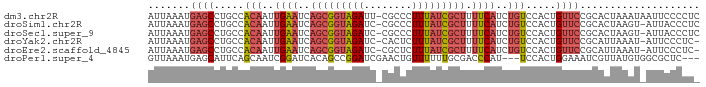
>dm3.chr2R 17302430 90 - 21146708 UUAAAUGAGCCUGCCACAAUUGAAUCAGCGGUAGAUU-CGCCCUUUAUCGCUUUUCAUCUGUCCACUGUUCCGCACUAAAUAAUUCCCCUC ......((((.....(((..((((..(((((((((..-.....))))))))).))))..))).....)))).................... ( -16.50, z-score = -2.12, R) >droSim1.chr2R 15956342 89 - 19596830 UUAAAUGAGCCUGCCACAAUUGAAUCAGCGGUAGAUC-CGCCCUUUAUCGCUUUUCAUCUGUCCACUGUUCCGCACUAAGU-AUUACCCUC ......((((.....(((..((((..(((((((((..-.....))))))))).))))..))).....))))..........-......... ( -16.50, z-score = -1.70, R) >droSec1.super_9 633395 89 - 3197100 UUAAAUGAGCCUGCCACAAUUGAAUCAGCGGUAGAUC-CGCCCUUUAUCGCUUUUCAUCUGUCCACUGUUCCGCACUAAGU-AUUACCCUC ......((((.....(((..((((..(((((((((..-.....))))))))).))))..))).....))))..........-......... ( -16.50, z-score = -1.70, R) >droYak2.chr2R 9254862 88 + 21139217 UUAAAUGAGCCUGCCACAAUUGAAUCAGCGGUAGAUC-CACUCUUUAUCGCUUUUCAUCUGUCCACUGUUCCGCAUUAAAU-AUUCCCUC- ......((((.....(((..((((..(((((((((..-.....))))))))).))))..))).....))))..........-........- ( -16.50, z-score = -2.44, R) >droEre2.scaffold_4845 11446918 88 - 22589142 UUAAAUGAGCCUGCCACAAUUGAAUCAGCGGUAGAUC-CGCUCUUUAUCGCUUUUCAUCUGUCCACUGUUCCGCAUUAAAU-AUUCCCUC- ......((((.....(((..((((..(((((((((..-.....))))))))).))))..))).....))))..........-........- ( -16.50, z-score = -1.99, R) >droPer1.super_4 3690255 85 + 7162766 UUAAAUGAGCAUUCAGCAAUCGGAUCACAGCCGGAUCGAACUGUUUUUUGCGACCCAU---UCCACUGGAAAUCGUUAUGUGGCGCUC--- ......((((((((.......))))....(((((.(((............))).)).(---(((...))))..........)))))))--- ( -15.50, z-score = 1.27, R) >consensus UUAAAUGAGCCUGCCACAAUUGAAUCAGCGGUAGAUC_CGCCCUUUAUCGCUUUUCAUCUGUCCACUGUUCCGCACUAAAU_AUUCCCCU_ ......((((.....(((..((((..(((((((((........))))))))).))))..))).....)))).................... (-10.61 = -11.92 + 1.31)



| Location | 17,302,430 – 17,302,521 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 80.68 |
| Shannon entropy | 0.36312 |
| G+C content | 0.44502 |
| Mean single sequence MFE | -16.33 |
| Consensus MFE | -10.61 |
| Energy contribution | -11.92 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.518798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17302430 91 - 21146708 AUUAAAUGAGCCUGCCACAAUUGAAUCAGCGGUAGAUU-CGCCCUUUAUCGCUUUUCAUCUGUCCACUGUUCCGCACUAAAUAAUUCCCCUC .......((((.....(((..((((..(((((((((..-.....))))))))).))))..))).....)))).................... ( -16.50, z-score = -2.13, R) >droSim1.chr2R 15956342 90 - 19596830 AUUAAAUGAGCCUGCCACAAUUGAAUCAGCGGUAGAUC-CGCCCUUUAUCGCUUUUCAUCUGUCCACUGUUCCGCACUAAGU-AUUACCCUC .......((((.....(((..((((..(((((((((..-.....))))))))).))))..))).....))))..........-......... ( -16.50, z-score = -1.68, R) >droSec1.super_9 633395 90 - 3197100 AUUAAAUGAGCCUGCCACAAUUGAAUCAGCGGUAGAUC-CGCCCUUUAUCGCUUUUCAUCUGUCCACUGUUCCGCACUAAGU-AUUACCCUC .......((((.....(((..((((..(((((((((..-.....))))))))).))))..))).....))))..........-......... ( -16.50, z-score = -1.68, R) >droYak2.chr2R 9254862 89 + 21139217 AUUAAAUGAGCCUGCCACAAUUGAAUCAGCGGUAGAUC-CACUCUUUAUCGCUUUUCAUCUGUCCACUGUUCCGCAUUAAAU-AUUCCCUC- .......((((.....(((..((((..(((((((((..-.....))))))))).))))..))).....))))..........-........- ( -16.50, z-score = -2.43, R) >droEre2.scaffold_4845 11446918 89 - 22589142 AUUAAAUGAGCCUGCCACAAUUGAAUCAGCGGUAGAUC-CGCUCUUUAUCGCUUUUCAUCUGUCCACUGUUCCGCAUUAAAU-AUUCCCUC- .......((((.....(((..((((..(((((((((..-.....))))))))).))))..))).....))))..........-........- ( -16.50, z-score = -1.99, R) >droPer1.super_4 3690255 86 + 7162766 GUUAAAUGAGCAUUCAGCAAUCGGAUCACAGCCGGAUCGAACUGUUUUUUGCGACCCAU---UCCACUGGAAAUCGUUAUGUGGCGCUC--- .......((((((((.......))))....(((((.(((............))).)).(---(((...))))..........)))))))--- ( -15.50, z-score = 1.51, R) >consensus AUUAAAUGAGCCUGCCACAAUUGAAUCAGCGGUAGAUC_CGCCCUUUAUCGCUUUUCAUCUGUCCACUGUUCCGCACUAAAU_AUUCCCCU_ .......((((.....(((..((((..(((((((((........))))))))).))))..))).....)))).................... (-10.61 = -11.92 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:45 2011