
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,300,502 – 17,300,596 |
| Length | 94 |
| Max. P | 0.996127 |

| Location | 17,300,502 – 17,300,596 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 75.58 |
| Shannon entropy | 0.45439 |
| G+C content | 0.49905 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -17.09 |
| Energy contribution | -18.28 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.996127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
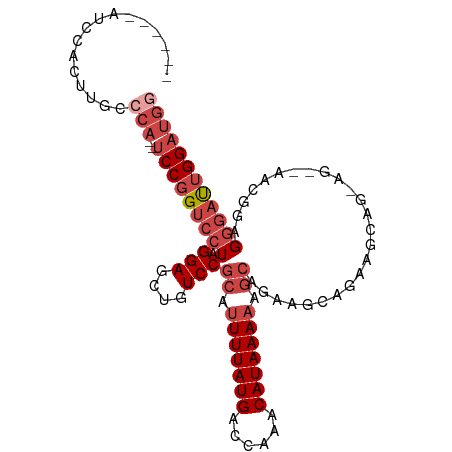
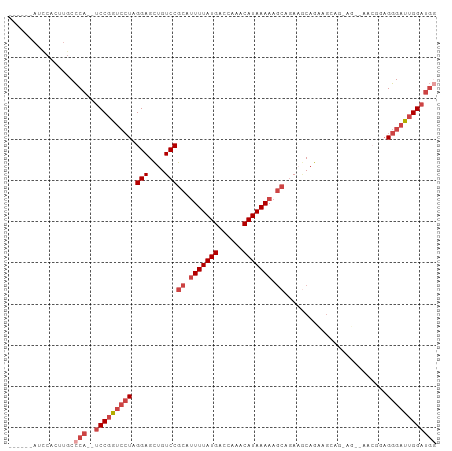
>dm3.chr2R 17300502 94 + 21146708 ------AGCCACUUGCCCA--UCCGGUCCUAGGAGCUGUCCGCAUUUUAUGACCAAACAUAAAAAGCAGAAGCAGCAGCAGAAG--AACGGAGGGAUUGGAUGU ------..(((....(((.--((((.((....))((((((.((.(((((((......))))))).)).)..)))))........--..)))))))..))).... ( -28.00, z-score = -1.63, R) >droPer1.super_4 3688393 85 - 7162766 --------------GCUCAAGUCCAGGGCUAGGAGUUGUCCGCAUUUUAUGGACAAACAUAAAAAA-AAACACAGACAGGAGGUUUGGGGCAGGGGCGGG---- --------------((((..((((.((((........))))...(((((((......)))))))..-.....(((((.....)))))))))..))))...---- ( -26.60, z-score = -3.27, R) >droEre2.scaffold_4845 11445028 88 + 22589142 ------AUCCACUUGCCCA--UCCGGUCCUAGGAGCUGUCCGCAUUUUAUGACCAAACAUAAAAAGCAGAUUCAGAGG--------AACGGAGGGAUUGGAUGG ------(((((....(((.--((((.((((..((...(((.((.(((((((......))))))).)).)))))..)))--------).)))))))..))))).. ( -33.00, z-score = -3.52, R) >droYak2.chr2R 9253043 94 - 21139217 AUCCACAACCACUUGCCCA--UCCGGUCGUAGGAGCUGUCCGCAGUUUAUGACCAAACAUAAAAAGCAGAAGCAGAAG--------AACGACGGGAUUGGAUGG ........(((....((.(--(((.(((((.....(((((.((..((((((......))))))..)).)..))))...--------.))))).)))).)).))) ( -27.70, z-score = -2.17, R) >droSec1.super_9 631493 94 + 3197100 ------AUCCACUUGCCCA--UCCGGUCCUAGGAGCUGUCCGCAUUUUAUGACCAAACAUAAAAAGCAGAAGCAGCAGCAGAAG--AACUGAGGGAUUGGAUGG ------..........(((--(((((((((....((((((.((.(((((((......))))))).)).)..)))))..(((...--..))).)))))))))))) ( -34.00, z-score = -3.59, R) >droSim1.chr2R 15954428 96 + 19596830 ------AUUCACUUGCCCA--UCCGGUCCUAGGAGCUGUCCGCAUUUUAUGACCAAACAUAAAAAGCAGAAGCAGCAGCAGCAGCAGAGGGAGGGAUUGGAUGG ------..........(((--(((((((((....((((((.((.(((((((......))))))).)).)..))))).((....)).......)))))))))))) ( -34.20, z-score = -2.77, R) >consensus ______AUCCACUUGCCCA__UCCGGUCCUAGGAGCUGUCCGCAUUUUAUGACCAAACAUAAAAAGCAGAAGCAGAAGCAG_AG__AACGGAGGGAUUGGAUGG .....................(((((((((.(((....)))((.(((((((......))))))).)).........................)))))))))... (-17.09 = -18.28 + 1.20)

| Location | 17,300,502 – 17,300,596 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 75.58 |
| Shannon entropy | 0.45439 |
| G+C content | 0.49905 |
| Mean single sequence MFE | -29.63 |
| Consensus MFE | -15.52 |
| Energy contribution | -16.68 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.982548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
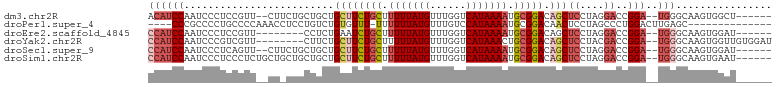
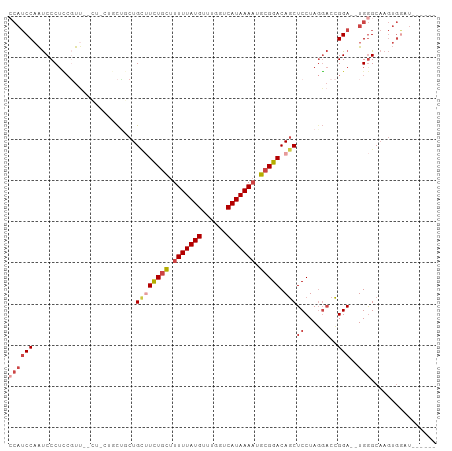
>dm3.chr2R 17300502 94 - 21146708 ACAUCCAAUCCCUCCGUU--CUUCUGCUGCUGCUUCUGCUUUUUAUGUUUGGUCAUAAAAUGCGGACAGCUCCUAGGACCGGA--UGGGCAAGUGGCU------ ....(((..(((((((.(--(((..(..((((..(((((.(((((((......))))))).)))))))))..).)))).))))--.)))....)))..------ ( -33.70, z-score = -3.11, R) >droPer1.super_4 3688393 85 + 7162766 ----CCCGCCCCUGCCCCAAACCUCCUGUCUGUGUUU-UUUUUUAUGUUUGUCCAUAAAAUGCGGACAACUCCUAGCCCUGGACUUGAGC-------------- ----..................(((.(((((((((((-.....((((......)))))))))))))))..(((.......)))...))).-------------- ( -14.40, z-score = -0.83, R) >droEre2.scaffold_4845 11445028 88 - 22589142 CCAUCCAAUCCCUCCGUU--------CCUCUGAAUCUGCUUUUUAUGUUUGGUCAUAAAAUGCGGACAGCUCCUAGGACCGGA--UGGGCAAGUGGAU------ ..(((((..(((((((.(--------((((((..(((((.(((((((......))))))).)))))))).....)))).))))--.)))....)))))------ ( -33.70, z-score = -3.75, R) >droYak2.chr2R 9253043 94 + 21139217 CCAUCCAAUCCCGUCGUU--------CUUCUGCUUCUGCUUUUUAUGUUUGGUCAUAAACUGCGGACAGCUCCUACGACCGGA--UGGGCAAGUGGUUGUGGAU ((((((.((((.(((((.--------.....((((((((..((((((......))))))..))))).)))....))))).)))--).))...))))........ ( -32.10, z-score = -2.72, R) >droSec1.super_9 631493 94 - 3197100 CCAUCCAAUCCCUCAGUU--CUUCUGCUGCUGCUUCUGCUUUUUAUGUUUGGUCAUAAAAUGCGGACAGCUCCUAGGACCGGA--UGGGCAAGUGGAU------ ..(((((..(((((.(((--((...(..((((..(((((.(((((((......))))))).)))))))))..).)))))..))--.)))....)))))------ ( -32.40, z-score = -2.44, R) >droSim1.chr2R 15954428 96 - 19596830 CCAUCCAAUCCCUCCCUCUGCUGCUGCUGCUGCUUCUGCUUUUUAUGUUUGGUCAUAAAAUGCGGACAGCUCCUAGGACCGGA--UGGGCAAGUGAAU------ ((((((..(((........((....)).((((..(((((.(((((((......))))))).))))))))).....)))..)))--)))..........------ ( -31.50, z-score = -2.13, R) >consensus CCAUCCAAUCCCUCCGUU__CU_CUGCUGCUGCUUCUGCUUUUUAUGUUUGGUCAUAAAAUGCGGACAGCUCCUAGGACCGGA__UGGGCAAGUGGAU______ ....(((..(((...................((((((((.(((((((......))))))).))))).)))(((.......)))...)))....)))........ (-15.52 = -16.68 + 1.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:42 2011