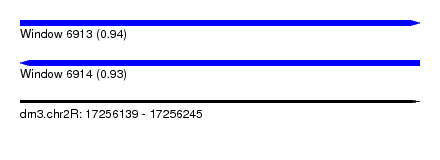
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,256,139 – 17,256,245 |
| Length | 106 |
| Max. P | 0.938648 |

| Location | 17,256,139 – 17,256,245 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.22 |
| Shannon entropy | 0.40200 |
| G+C content | 0.49231 |
| Mean single sequence MFE | -32.42 |
| Consensus MFE | -20.53 |
| Energy contribution | -20.73 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.938648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
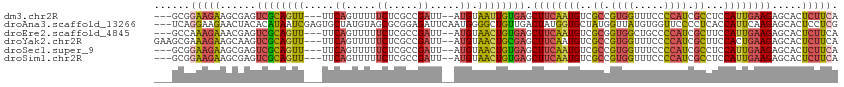
>dm3.chr2R 17256139 106 + 21146708 ---GCGGAAGAAGCGAGUCGCAGUU---UUCAGUUUUUCUCGCCGAUU--AUGUAAUUGUGAGCUUCAAUGUCGCCGUGGUUUCCCCAUCGCCUCCAUUGAAGAGCACUCUUCA ---...(((((.(((((..((....---....))....))))).....--........(((..((((((((..((.((((.....)))).))...))))))))..)))))))). ( -32.60, z-score = -2.53, R) >droAna3.scaffold_13266 5062199 111 + 19884421 ---UCAGGAAGAACUACACAUAAUCGAGUGCUAUGUAGCGCGGAGAUUCAAUGGGGCUGUUGACUAUGGUGCUAUGGUUAUGUGGUUCCCCUCACCAUUCAAGAGCACUCCUCG ---..((((.((((..(((((((((..(((((....))))).........((((.(((((.....))))).)))))))))))))))))..(((.........)))...)))).. ( -30.00, z-score = 0.15, R) >droEre2.scaffold_4845 11400568 106 + 22589142 ---GCCAAAGAAACGAGUCGCAGUU---UUCAGUUUUUCUCGCCGAUU--AUGUAACUGUGAGCUUCAAUGUCGCGGUGGCUGCCCCAUCGCUUCCAUUGAAGAGCACUCUUCA ---......(((..(((((((((((---..((.....((.....))..--.)).)))))))).((((((((..(((((((.....)))))))...))))))))....)))))). ( -32.20, z-score = -2.09, R) >droYak2.chr2R 9207953 109 - 21139217 GAAGCGAAAGAAGCAAGUCGCAGUU---UUCAGUUUUUCUCGCCGAUU--AUGUAACUGCGAGCUUCAAUGUCGCCGUGGUUUCCCCAUCGCUUCCACUGAAGAGCACUCUUCA (((((((..(((((...((((((((---..((.....((.....))..--.)).)))))))))))))..........(((.....))))))))))....(((((....))))). ( -34.50, z-score = -2.86, R) >droSec1.super_9 587433 106 + 3197100 ---GCGGAAGAAGCGAGUCGCAGUU---UUCAGUUUUUCUCGCCGAUU--AUGUAACUGUGAGCUUCAAUGUCGCCGUGGUUUCCCCAUCGCCUCCAUUGAAGAGCACUCUUCA ---...(((((.(((((..((....---....))....))))).....--........(((..((((((((..((.((((.....)))).))...))))))))..)))))))). ( -32.60, z-score = -2.33, R) >droSim1.chr2R 15909055 106 + 19596830 ---GCGGAAGAAGCGAGUCGCAGUU---UUCAGUUUUUCUCGCCGAUU--AUGUAACUGUGAGCUUCAAUGUCGCCGUGGUUUCCCCAUCGCCUCCAUUGAAGAGCACUCUUCA ---...(((((.(((((..((....---....))....))))).....--........(((..((((((((..((.((((.....)))).))...))))))))..)))))))). ( -32.60, z-score = -2.33, R) >consensus ___GCGGAAGAAGCGAGUCGCAGUU___UUCAGUUUUUCUCGCCGAUU__AUGUAACUGUGAGCUUCAAUGUCGCCGUGGUUUCCCCAUCGCCUCCAUUGAAGAGCACUCUUCA ......(((((......((((((((......((((.........))))......)))))))).((((((((..((.((((.....)))).))...)))))))).....))))). (-20.53 = -20.73 + 0.20)
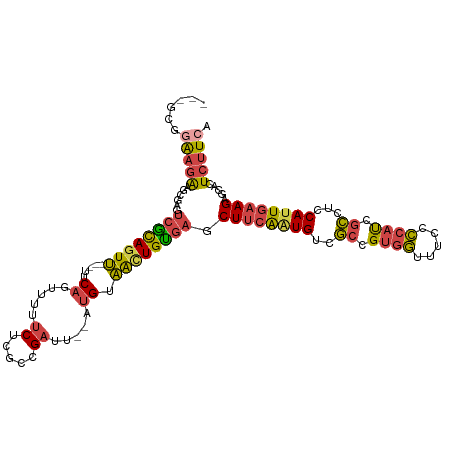
| Location | 17,256,139 – 17,256,245 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.22 |
| Shannon entropy | 0.40200 |
| G+C content | 0.49231 |
| Mean single sequence MFE | -33.06 |
| Consensus MFE | -20.07 |
| Energy contribution | -21.05 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17256139 106 - 21146708 UGAAGAGUGCUCUUCAAUGGAGGCGAUGGGGAAACCACGGCGACAUUGAAGCUCACAAUUACAU--AAUCGGCGAGAAAAACUGAA---AACUGCGACUCGCUUCUUCCGC--- .((((((((..((((((((..(.((...((....)).)).)..))))))))..)))........--....((((((..........---........)))))))))))...--- ( -35.17, z-score = -3.40, R) >droAna3.scaffold_13266 5062199 111 - 19884421 CGAGGAGUGCUCUUGAAUGGUGAGGGGAACCACAUAACCAUAGCACCAUAGUCAACAGCCCCAUUGAAUCUCCGCGCUACAUAGCACUCGAUUAUGUGUAGUUCUUCCUGA--- ..(((((.(((.((((((((((..((...........))....))))))..)))).)))..............(.((((((((...........)))))))).))))))..--- ( -27.40, z-score = 0.02, R) >droEre2.scaffold_4845 11400568 106 - 22589142 UGAAGAGUGCUCUUCAAUGGAAGCGAUGGGGCAGCCACCGCGACAUUGAAGCUCACAGUUACAU--AAUCGGCGAGAAAAACUGAA---AACUGCGACUCGUUUCUUUGGC--- .((((((((..((((((((...(((.(((.....))).)))..))))))))..)))........--....((((((..........---........)))))))))))...--- ( -29.27, z-score = -0.65, R) >droYak2.chr2R 9207953 109 + 21139217 UGAAGAGUGCUCUUCAGUGGAAGCGAUGGGGAAACCACGGCGACAUUGAAGCUCGCAGUUACAU--AAUCGGCGAGAAAAACUGAA---AACUGCGACUUGCUUCUUUCGCUUC ((((((....))))))...(((((((..((....))...........(((((((((((((....--..((((.........)))).---))))))))...)))))..))))))) ( -36.20, z-score = -2.38, R) >droSec1.super_9 587433 106 - 3197100 UGAAGAGUGCUCUUCAAUGGAGGCGAUGGGGAAACCACGGCGACAUUGAAGCUCACAGUUACAU--AAUCGGCGAGAAAAACUGAA---AACUGCGACUCGCUUCUUCCGC--- .((((((((..((((((((..(.((...((....)).)).)..))))))))..)))........--....((((((..........---........)))))))))))...--- ( -35.17, z-score = -2.85, R) >droSim1.chr2R 15909055 106 - 19596830 UGAAGAGUGCUCUUCAAUGGAGGCGAUGGGGAAACCACGGCGACAUUGAAGCUCACAGUUACAU--AAUCGGCGAGAAAAACUGAA---AACUGCGACUCGCUUCUUCCGC--- .((((((((..((((((((..(.((...((....)).)).)..))))))))..)))........--....((((((..........---........)))))))))))...--- ( -35.17, z-score = -2.85, R) >consensus UGAAGAGUGCUCUUCAAUGGAGGCGAUGGGGAAACCACGGCGACAUUGAAGCUCACAGUUACAU__AAUCGGCGAGAAAAACUGAA___AACUGCGACUCGCUUCUUCCGC___ .((((((((..((((((((...((..(((.....)))..))..))))))))..)))..............((((((.....................)))))))))))...... (-20.07 = -21.05 + 0.98)

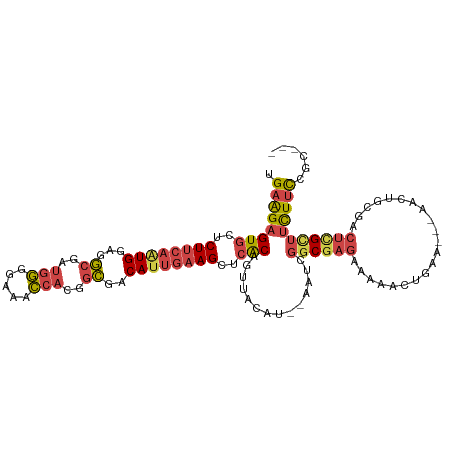
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:39 2011