
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,254,978 – 17,255,078 |
| Length | 100 |
| Max. P | 0.836774 |

| Location | 17,254,978 – 17,255,078 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 70.80 |
| Shannon entropy | 0.55084 |
| G+C content | 0.48721 |
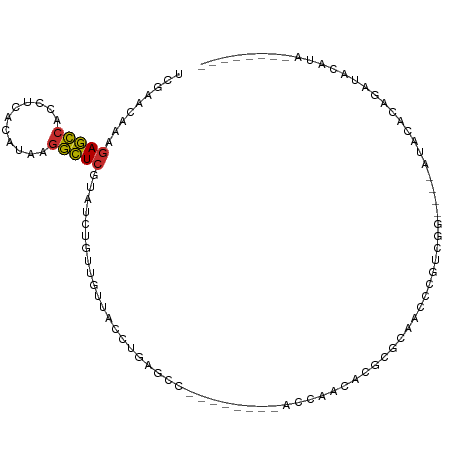
| Mean single sequence MFE | -20.15 |
| Consensus MFE | -6.66 |
| Energy contribution | -6.49 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.836774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
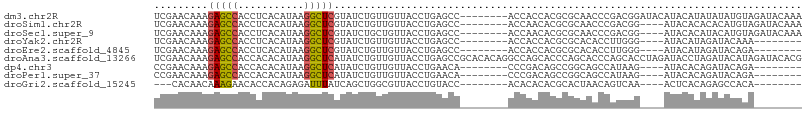
>dm3.chr2R 17254978 100 - 21146708 UCGAACAAAGAGCCACCUCACAUAAGGCUCGUAUCUGUUGUUACCUGAGCC--------ACCACCACGCGCAACCCGACGGAUACAUACAUAUAUAUGUAGAUACAAA .........(((((...........)))))((((((((((.....((.((.--------........)).))...)))))))))).(((((....)))))........ ( -23.70, z-score = -2.90, R) >droSim1.chr2R 15907919 96 - 19596830 UCGAACAAAGAGCCACCUCACAUAAGGCUCGUAUCUGUUGUUACCUGAGCC--------ACCAACACGCGCAACCCGACGG----AUACACACACAUGUAGAUACAAA ((..(((..(((((...........)))))((((((((((.....((.((.--------........)).))...))))))----)))).......))).))...... ( -22.50, z-score = -2.56, R) >droSec1.super_9 586306 96 - 3197100 UCGAACAAAGAGCCACCUCACAUAAGGCUCGUAUCUGCUGUUACCUGAGCC--------ACCAACACGCGCAACCCGACGG----AUACACAUACAUGUAGAUACAAA .........(((((...........)))))(((((((((((....((.(((--------........).))..((....))----...))...))).))))))))... ( -21.80, z-score = -2.40, R) >droYak2.chr2R 9206756 88 + 21139217 UCGAACAAAGAGCCACCUCACAUAAGGCUCGUAUCUGUUGUUACCUGAGCC--------ACCACCACGCGCACACCUUGGG----AUACAUAGAUACAAA-------- .........(((((...........)))))((((((((.((..((..((..--------................))..))----..))))))))))...-------- ( -22.17, z-score = -2.63, R) >droEre2.scaffold_4845 11399441 88 - 22589142 UCGAACAAAGAGCCACCUCACAUAAGGCUCGUAUCUGUUGUUACCUGAGCC--------ACCACCACGCGCACACCUUGGG----AUACAUAGAUACAGA-------- .........(((((...........)))))((((((((.((..((..((..--------................))..))----..))))))))))...-------- ( -22.17, z-score = -2.23, R) >droAna3.scaffold_13266 5061156 108 - 19884421 UCGAACAAAGAGCCACCACACAUAAGGCUCAUAUCUGUUGUUACCUGAGCCGCACACAGGCCAGCACCCAGCACCCAGCACCUAGAUACCUAGAUACAUAGAUACACG .........(((((...........))))).(((((((((....(((.(((.......))))))....)))).........((((....))))......))))).... ( -25.10, z-score = -3.57, R) >dp4.chr3 368915 88 + 19779522 CCGAACAAAGAGCCACCACACAUAAGGCUCAUAUCUGUUGUUACCUGAACA--------CCCGACAGCCGGCAGCCAUAAG----AUACACAGAUACAGA-------- .........(((((...........))))).(((((((.((...((.....--------.(((.....)))........))----..)))))))))....-------- ( -17.94, z-score = -2.59, R) >droPer1.super_37 349440 88 + 760358 CCGAACAAAGAGCCACCACACAUAAGGCUCAUAUCUGUUGUUACCUGAACA--------CCCGACAGCCGGCAGCCAUAAG----AUACACAGAUACAGA-------- .........(((((...........))))).(((((((.((...((.....--------.(((.....)))........))----..)))))))))....-------- ( -17.94, z-score = -2.59, R) >droGri2.scaffold_15245 14174494 85 + 18325388 ---CACAACAAAGAACACCACAGAGAUUUAUCAGCUGGCGUUACCUGUACC--------ACACACACGCACUAACAGUCAA----ACUCACAGAGCCACA-------- ---....(((...(((.((((.((......)).).))).)))...)))...--------........((.((...((....----.))...)).))....-------- ( -8.00, z-score = 0.68, R) >consensus UCGAACAAAGAGCCACCUCACAUAAGGCUCGUAUCUGUUGUUACCUGAGCC________ACCAACACGCGCAACCCGUCGG____AUACACAGAUACAUA________ .........(((((...........))))).............................................................................. ( -6.66 = -6.49 + -0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:37 2011