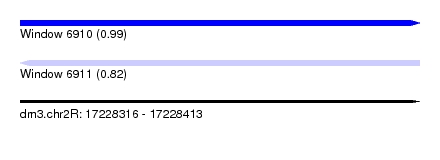
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,228,316 – 17,228,413 |
| Length | 97 |
| Max. P | 0.994151 |

| Location | 17,228,316 – 17,228,413 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.43 |
| Shannon entropy | 0.42771 |
| G+C content | 0.57830 |
| Mean single sequence MFE | -41.30 |
| Consensus MFE | -28.09 |
| Energy contribution | -29.87 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.994151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
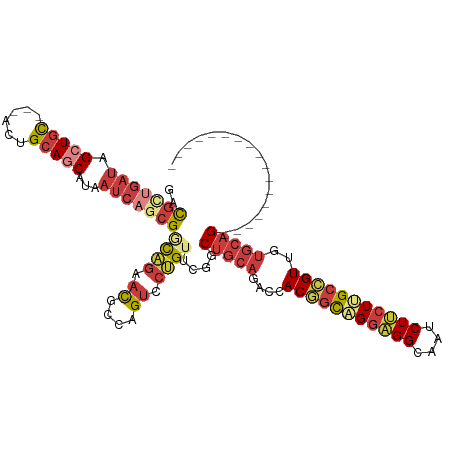
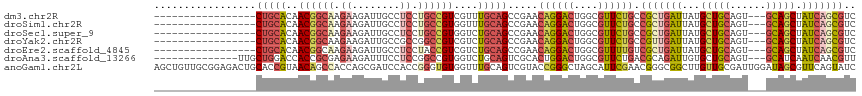
>dm3.chr2R 17228316 97 + 21146708 GACGCUGAUAGCUGC---ACUGCAGCAUAAUCAGCGGCAGAACGCCAGUCCUGUUCGGCUGCAAACGACGGCAGGAGGCAAUCUUCUUGCCGUUGUGCAG----------------- (((((((((.(((((---...)))))...))))))(((.....))).)))........((((..((((((((((((((....))))))))))))))))))----------------- ( -47.80, z-score = -4.43, R) >droSim1.chr2R 15881035 97 + 19596830 GACGCUGAUAGCUGC---ACUGCAGCAUAAUCAGCGGCAGAACGCCAGUCCUGUUCGGCUGCAAACCACGGCAGGAGGCAAUCUUCUUGCCGUUGUGCAG----------------- (((((((((.(((((---...)))))...))))))(((.....))).)))........(((((....(((((((((((....)))))))))))..)))))----------------- ( -45.00, z-score = -3.72, R) >droSec1.super_9 559668 97 + 3197100 GACGCUGAUAGCUGC---ACUGCAGCAUAAUCAGCGGCAGAACGCCAGUCCUGUUCGGCUGCAGACCACGGCAGGAGGCAAUCUUCUUGCCGUUGUGCAG----------------- (((((((((.(((((---...)))))...))))))(((.....))).)))........(((((.(..(((((((((((....)))))))))))).)))))----------------- ( -44.90, z-score = -3.32, R) >droYak2.chr2R 9179340 97 - 21139217 GACGCUGAUAGCUGC---ACUGCAGCAUAAUCAACGGCAGAACGCCAGUCCUGUUCGGCUGCAGACGACGGCCGGCGGCAAUCUUCUUGCCGUUGUGCAG----------------- ((((.((((.(((((---...)))))...)))).)(((.....))).)))((((.(((((((....).)))))((((((((.....))))))))).))))----------------- ( -36.70, z-score = -0.93, R) >droEre2.scaffold_4845 11372397 97 + 22589142 GACGCUGAUAGCUGC---ACUGCAGCAUAAUCAGCGACAAAACGCCAGUCCUGUUCGGCUGCAGACGACGGUAGGAGGCAAUCUUCUUGCCGUUGUGCAG----------------- ...........((((---.(((((((.....(((.(((.........))))))....)))))))((((((((((((((....))))))))))))))))))----------------- ( -43.60, z-score = -3.91, R) >droAna3.scaffold_13266 5030806 100 + 19884421 AACGUUGAUUGAUGC---ACUGCAGCACAAUCUGCGUCAGAACGCCAGUCCAGUGCGACUGCAGACCACGGCCGGAGGAAAUCUUCUCGCGGUGGUCCAGCAA-------------- .((((.(((((.(((---......)))))))).))))......(((((((......)))))..((((((.((.(((((....))))).)).))))))..))..-------------- ( -39.80, z-score = -2.20, R) >anoGam1.chr2L 26897997 117 + 48795086 GAUACUGAACGCUAUCCAAUCGCAACAAGCCGCCCGUUCGAAUGCUAGCCCGGUACGACUGCAAACCACACCCGGUGGAUCGCUGGUGGCUGUUACGGUGCAGUCUCCGCAACAGCU ..(((((...((((..((.(((.(((.........)))))).)).)))).))))).....((...((((.....))))...((.((.((((((......)))))).))))....)). ( -31.30, z-score = 1.23, R) >consensus GACGCUGAUAGCUGC___ACUGCAGCAUAAUCAGCGGCAGAACGCCAGUCCUGUUCGGCUGCAGACCACGGCAGGAGGCAAUCUUCUUGCCGUUGUGCAG_________________ ..(((((((.(((((......)))))...)))))))(((.(((((.((((......)))))).......(((((((((....)))))))))))).)))................... (-28.09 = -29.87 + 1.78)

| Location | 17,228,316 – 17,228,413 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.43 |
| Shannon entropy | 0.42771 |
| G+C content | 0.57830 |
| Mean single sequence MFE | -38.96 |
| Consensus MFE | -25.84 |
| Energy contribution | -27.26 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.818168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17228316 97 - 21146708 -----------------CUGCACAACGGCAAGAAGAUUGCCUCCUGCCGUCGUUUGCAGCCGAACAGGACUGGCGUUCUGCCGCUGAUUAUGCUGCAGU---GCAGCUAUCAGCGUC -----------------..(((.((((((((.....)))))....(((((((((((....)))))..))).)))))).)))(((((((...(((((...---))))).))))))).. ( -39.20, z-score = -2.02, R) >droSim1.chr2R 15881035 97 - 19596830 -----------------CUGCACAACGGCAAGAAGAUUGCCUCCUGCCGUGGUUUGCAGCCGAACAGGACUGGCGUUCUGCCGCUGAUUAUGCUGCAGU---GCAGCUAUCAGCGUC -----------------(((((..((((((.((........)).))))))....))))).....((((((....)))))).(((((((...(((((...---))))).))))))).. ( -40.00, z-score = -1.97, R) >droSec1.super_9 559668 97 - 3197100 -----------------CUGCACAACGGCAAGAAGAUUGCCUCCUGCCGUGGUCUGCAGCCGAACAGGACUGGCGUUCUGCCGCUGAUUAUGCUGCAGU---GCAGCUAUCAGCGUC -----------------(((((..((((((.((........)).))))))....))))).....((((((....)))))).(((((((...(((((...---))))).))))))).. ( -40.00, z-score = -1.79, R) >droYak2.chr2R 9179340 97 + 21139217 -----------------CUGCACAACGGCAAGAAGAUUGCCGCCGGCCGUCGUCUGCAGCCGAACAGGACUGGCGUUCUGCCGUUGAUUAUGCUGCAGU---GCAGCUAUCAGCGUC -----------------..(((.((((((((.....)))))(((((((.(((.(....).)))...)).)))))))).)))(((((((...(((((...---))))).))))))).. ( -42.00, z-score = -2.30, R) >droEre2.scaffold_4845 11372397 97 - 22589142 -----------------CUGCACAACGGCAAGAAGAUUGCCUCCUACCGUCGUCUGCAGCCGAACAGGACUGGCGUUUUGUCGCUGAUUAUGCUGCAGU---GCAGCUAUCAGCGUC -----------------..(((.((((((((.....))))).......(((((((...........)))).)))))).)))(((((((...(((((...---))))).))))))).. ( -32.90, z-score = -0.73, R) >droAna3.scaffold_13266 5030806 100 - 19884421 --------------UUGCUGGACCACCGCGAGAAGAUUUCCUCCGGCCGUGGUCUGCAGUCGCACUGGACUGGCGUUCUGACGCAGAUUGUGCUGCAGU---GCAUCAAUCAACGUU --------------.....(((((((.(((((........)))..)).))))))).(((((......)))))((((....)))).((((((((......---))).)))))...... ( -35.30, z-score = -0.53, R) >anoGam1.chr2L 26897997 117 - 48795086 AGCUGUUGCGGAGACUGCACCGUAACAGCCACCAGCGAUCCACCGGGUGUGGUUUGCAGUCGUACCGGGCUAGCAUUCGAACGGGCGGCUUGUUGCGAUUGGAUAGCGUUCAGUAUC .((((((((((........))))))))))..((.((((.((((.....)))).))))(((((((.((((((.((..........)))))))).)))))))))............... ( -43.30, z-score = -0.59, R) >consensus _________________CUGCACAACGGCAAGAAGAUUGCCUCCUGCCGUGGUCUGCAGCCGAACAGGACUGGCGUUCUGCCGCUGAUUAUGCUGCAGU___GCAGCUAUCAGCGUC .................(((((..((((((.((........)).))))))....))))).....((((((....)))))).(((((((...(((((......))))).))))))).. (-25.84 = -27.26 + 1.42)
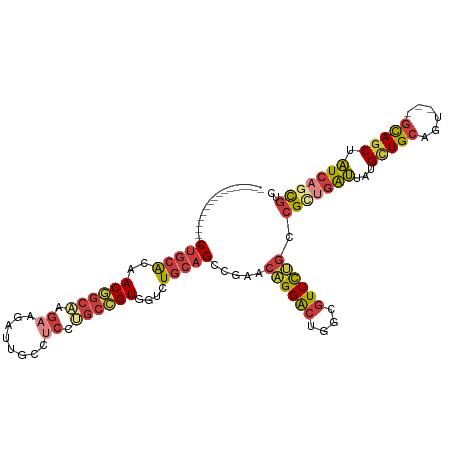
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:36 2011