
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,221,200 – 17,221,300 |
| Length | 100 |
| Max. P | 0.531358 |

| Location | 17,221,200 – 17,221,300 |
|---|---|
| Length | 100 |
| Sequences | 15 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 68.43 |
| Shannon entropy | 0.71818 |
| G+C content | 0.55668 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -11.55 |
| Energy contribution | -11.31 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.79 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.531358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
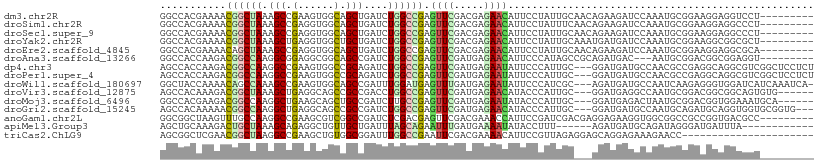
>dm3.chr2R 17221200 100 + 21146708 GGCCACGAAAACGGCUAAAGCCGAAGUGGCAGCUGAUCUGGCCGAGUUCGACGAGAACAUUCCUAUUGCAACAGAAGAUCCAAAUGCGGAAGGAGGUCCU--------- .(((((.....((((....))))..))))).........((((..((((.....))))..((((.(((((..............))))).))))))))..--------- ( -32.14, z-score = -1.63, R) >droSim1.chr2R 15873421 100 + 19596830 GGCCACGAAAACGGCUAAAGCCGAGGUGGCAGCUGAUCUGGCCGAGUUCGACGAGAACAUUCCUAUUUCAACAGAAGAUCCAAAUGCGGAAGGAGGCCCU--------- .(((((.....((((....))))..))))).........((((..((((.....))))..((((.((((....)))).(((......)))))))))))..--------- ( -33.30, z-score = -1.68, R) >droSec1.super_9 552590 100 + 3197100 GGCCACGAAAACGGCUAAAGCCGAGGUGGCAGCUGAUCUGGCCGAGUUCGACGAGAACAUUCCUAUUGCAACAGAAGAUCCAAAUGCGGAAGGAGGCCCU--------- .(((((.....((((....))))..))))).........((((..((((.....))))..((((.(((((..............))))).))))))))..--------- ( -33.74, z-score = -1.71, R) >droYak2.chr2R 9172034 100 - 21139217 GGCCACGAAAACGGCUAAAGCUGAGGUGGCUGCUGAUCUGGCCGAGUUCGACGAGAACAUUCCUAUUGCAAAUGAUGAUCCAAAUGCGGAAGGCGGCGCU--------- ((((((.....((((....))))..)))))).........((((.((((.....))))...(((.(((((..((......))..))))).)))))))...--------- ( -31.10, z-score = -0.64, R) >droEre2.scaffold_4845 11364985 100 + 22589142 GGCCACGAAAACAGCUAAAGCCGAGGUGGCAGCUGAUCUGGCCGAGUUCGACGAGAACAUUCCUAUUGCAACAGAAGAUCCAAAUGCGGAAGGAGGCGCA--------- (((((.((...(((((...(((.....)))))))).)))))))..((((.....)))).(((((.(((((..............))))).))))).....--------- ( -31.44, z-score = -1.32, R) >droAna3.scaffold_13266 5023828 97 + 19884421 GGCCACCAAGACGGCCAAGGCGGAGGCGGCAGCCGAUCUGGCCGAGUUCGAUGAGAACAUUCCCAUAGCCGCAGAUGAC---AAUGCGGACGGCGGAGGU--------- .(((.((.(((((((...(.((....)).).)))).))).((((.((((.....))))..........(((((......---..))))).)))))).)))--------- ( -35.90, z-score = -0.90, R) >dp4.chr3 9987378 106 - 19779522 AGCCACCAAGACGGCCAAGGCCGAAGUGGCCGCAGAUCUGGCCGAGUUCGAUGAGAAUAUUCCCAUUGC---GGAUGAUGCCAACGCCGAGGCAGGCGUCGGCUCCUCU .(((((.....((((....))))..)))))(((((...(((....((((.....))))....)))))))---)((.((.(((.(((((......))))).))))).)). ( -41.20, z-score = -1.24, R) >droPer1.super_4 2055185 106 - 7162766 AGCCACCAAGACGGCCAAGGCCGAAGUGGCCGCAGAUCUGGCCGAGUUCGAUGAGAAUAUUCCCAUUGC---GGAUGAUGCCAACGCCGAGGCAGGCGUCGGCUCCUCU .(((((.....((((....))))..)))))(((((...(((....((((.....))))....)))))))---)((.((.(((.(((((......))))).))))).)). ( -41.20, z-score = -1.24, R) >droWil1.scaffold_180697 1476052 105 + 4168966 GGCUACCAAAACAGCCAAAGCCGAAGUGGCAGCCGAUUUGGAUGAGUUUGAUGAGAAUAUUCCCAUCGC---AGAUGAUGCCAAUCAAGAGGGUGGAUCAUCAAAUCA- ((((.(((...(.((....)).)...))).))))(((((..........((((.((....)).))))..---.((((((.(((..(....)..)))))))))))))).- ( -29.90, z-score = -1.34, R) >droVir3.scaffold_12875 2552812 100 - 20611582 AGCCACAAAGACGGCUAAAGCUGAGGCAGCCGCCGACCUGGCCGAGUUCGAUGAGAACAUACCCAUUGC---GGAUGAGGCCAAUGCGGACGGCGGCAGUGUG------ ...((((....((((....)))).....(((((((.(((((((..((((((((.(......).)))..)---))))..)))))....)).)))))))..))))------ ( -39.70, z-score = -2.37, R) >droMoj3.scaffold_6496 15187026 100 - 26866924 GGCCACGAAGACGGCCAAGGCUGAAGCAGCUGCCGAUCUUGCCGAGUUCGAUGAGAAUAUACCCAUUGC---GGAUGAGACUAAUGCGGACGGUGGAAAUGCA------ ((((........)))).........(((.(..(((.((((..((....))..)))).....(((((((.---(.......)))))).)).)))..)...))).------ ( -30.20, z-score = -0.90, R) >droGri2.scaffold_15245 14137868 103 - 18325388 AGCCACAAAAACGGCCAAGGCUGAGGCAGCCGCCGAUCUGGCCGAGUUCGAUGAGAACAUACCCAUUGC---GGAUGAUGCCAAUGCAGAUGCAGGUGGUGCGGUG--- .(((.......((((....))))..(((.(((((....((((...((((.....))))....((.....---)).....)))).(((....)))))))))))))).--- ( -35.20, z-score = -0.49, R) >anoGam1.chr2L 26886788 100 + 48795086 GGCGGCUAAGUUUGCCAAGGCCGAAGCGUCGGCCGAUCUCGACGAGUUCGACGAAACCAUUCCGAUCGACGAGGAGAAGGUGGCGGCCGCCGGUGACGCC--------- (((((((..(.(..((..((((((....))))))..(((((.(((..(((..(((....))))))))).)))))....))..)))))))))(....)...--------- ( -41.90, z-score = -0.86, R) >apiMel3.Group3 6963370 91 + 12341916 AGCUGCAAAGACUGCUAAAGCAGAGGCUGUUGCUGAUUUAGCAGAAUUUGAUGAAAAUAUACCUUU------AGAUGAUGCAGAUAGGGAUGAUUUA------------ ..(((((....((((((((.....(((....)))..)))))))).((((((.............))------))))..)))))..............------------ ( -20.52, z-score = -0.94, R) >triCas2.ChLG9 9475384 87 - 15222296 AGCGGCUCGAACGGCUAAGGCCGAAGCUGUGGCGGAUUUGGCCGAAUUCGACGAAAACAUUCCGUUAGAGGAGCAGGAGAAAGAACC---------------------- ....((((...((((....))))...((.(((((((((((..((....)).)))).....))))))).)))))).............---------------------- ( -26.60, z-score = -0.65, R) >consensus GGCCACGAAAACGGCUAAAGCCGAGGUGGCAGCCGAUCUGGCCGAGUUCGAUGAGAACAUUCCCAUUGC___AGAUGAUGCAAAUGCGGAAGGAGGCGCU_________ ...........((((((.(((.(......).)))....)))))).((((.....))))................................................... (-11.55 = -11.31 + -0.23)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:33 2011