| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,528,948 – 3,529,038 |
| Length | 90 |
| Max. P | 0.933513 |

| Location | 3,528,948 – 3,529,038 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 60.60 |
| Shannon entropy | 0.80028 |
| G+C content | 0.50589 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -12.73 |
| Energy contribution | -12.73 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.65 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
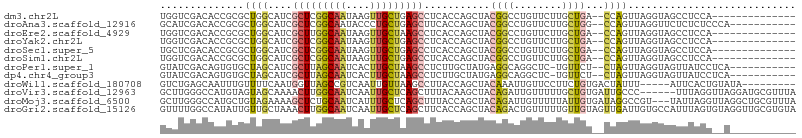
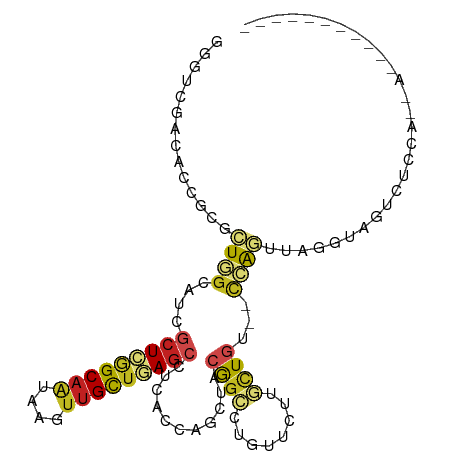
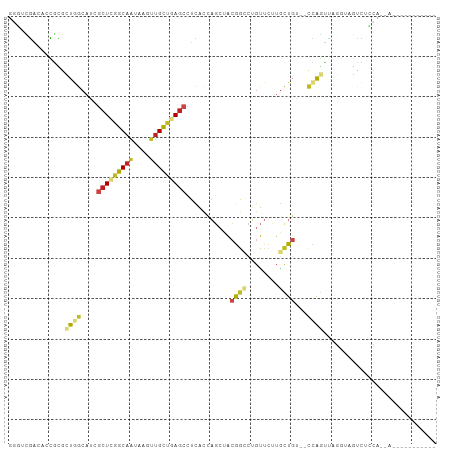
>dm3.chr2L 3528948 90 - 23011544 UGGUCGACACCGCGCUGGCAUCGCUCGGCAAUAAGUUGCUGAGCCUCACCAGCUACGGCCUGUUCUUGCUGA--CCAGUUAGGUAGCCUCCA-------------- .((..(..(((..(((((....(((((((((....)))))))))....)))))..((((........)))).--.......)))..)..)).-------------- ( -33.80, z-score = -1.37, R) >droAna3.scaffold_12916 5576053 93 - 16180835 GCAUCGACACCGCGCUGGCAUCGCUCGGCAAUACCCUGCUGAGCUUCACCAGCUACGGCCUGUUCUUGCUGG--CCAGUUAGGUUCUCUCUCCCA----------- .....((.(((..(((((....((((((((......))))))))....)))))...((((.((....)).))--)).....))))).........----------- ( -35.20, z-score = -2.92, R) >droEre2.scaffold_4929 3567296 90 - 26641161 UGGUCGACACCGCGCUGGCAUCGCUUGGCAAUAAGUUGCUAAGCCUCACCAGCUACGGCCUGUUCUUGCUGA--CCAGUUAGGUAGCCUCCA-------------- .((..(..(((..(((((....(((((((((....)))))))))....)))))..((((........)))).--.......)))..)..)).-------------- ( -31.90, z-score = -1.17, R) >droYak2.chr2L 3519061 90 - 22324452 UGGUCGACACCGCGCUGGCAUCGCUCGGCAAUAAGUUGCUGAGCCUCACCAGCUACGGCCUGUUCUUGCUGA--CCAGUUAGGUAGCCUCCA-------------- .((..(..(((..(((((....(((((((((....)))))))))....)))))..((((........)))).--.......)))..)..)).-------------- ( -33.80, z-score = -1.37, R) >droSec1.super_5 1631288 90 - 5866729 UGCUCGACACCGCGCUGGCAUCGCUCGGCAAUAAGUUGCUGAGCCUCACCAGCUACGGCCUGUUCUUGCUGA--CCAGUUAGGUAGCCUCCA-------------- ((((..((.....(((((....(((((((((....)))))))))....)))))..((((........)))).--...))..)))).......-------------- ( -32.50, z-score = -1.20, R) >droSim1.chr2L 3484046 90 - 22036055 UGGUCGACACCGCGCUGGCAUCGCUCGGCAAUAAGUUGCUGAGCCUCACCAGCUACGGCCUGUUCUUGCUGA--CCAGUUAGGUAGCCUCCA-------------- .((..(..(((..(((((....(((((((((....)))))))))....)))))..((((........)))).--.......)))..)..)).-------------- ( -33.80, z-score = -1.37, R) >droPer1.super_1 3571904 92 + 10282868 GUAUCGACAGUGUGCUAGCAUCGCUUAGCAAUCACUUGCUAAGCCUCUUGCUAUGAGGCAGGCUC-UGUUCU--CUAGUUAGGUAGUUAUCCUCA----------- .....(((((...(((.((...(((((((((....)))))))))(((.......))))).))).)-))))..--......(((.......)))..----------- ( -26.20, z-score = -0.63, R) >dp4.chr4_group3 6472411 92 + 11692001 GUAUCGACAGUGUGCUAGCAUCGCUUAGCAAUCACUUGCUAAGCCUCUUGCUAUGAGGCAGGCUC-UGUUCU--CUAGUUAGGUAGUUAUCCUCA----------- .....(((((...(((.((...(((((((((....)))))))))(((.......))))).))).)-))))..--......(((.......)))..----------- ( -26.20, z-score = -0.63, R) >droWil1.scaffold_180708 9008697 92 - 12563649 GUCUGAGCAAUUUGUUUUCAAUGGUUAGCCGUCAAUUGUUAAGCCUUACCAGCUACAAAUUGUUCCUUCUGUGACUAUUU-----AUUCACUGUAUA--------- ....(((((((((((......((((.((.(............).)).))))...))))))))))).....((((......-----..))))......--------- ( -18.50, z-score = -1.89, R) >droVir3.scaffold_12963 4958514 100 + 20206255 GCUUGGGCCAUGUAGUAGCAAAACUUGGCAAUCAAUUGCUCAGCUUUACAAGCUACAGAUUGUUUUUGCUGUGAUUGCCC------UUUAGGUUAGGAUGCGUUUA ((...((((.(((....)))......((((((((......((((...((((........))))....)))))))))))).------....)))).....))..... ( -24.10, z-score = 0.77, R) >droMoj3.scaffold_6500 14125061 103 + 32352404 GCUUGGGCCAUGCUGUAGAAAAGCUCUGCAAUCAUUUGCUCAGCUUUACCAGCUACAGAUUGUUUUUAUUGUGAUAGGCCGU---UAUUAGGUUAGGCUGCGUUUA ((...((((...((((((.((((((..((((....))))..)))))).....))))))..................((((..---.....)))).))))))..... ( -28.20, z-score = -0.16, R) >droGri2.scaffold_15126 4159687 106 - 8399593 GUUUUGGCCAUAUUGUUGCUAAACUUGGCAAUCAAUUGCUCAGCUUCACCAGCUACAGACUGUUUUUGUUGUAGUUGAUUGUGCCAUUUAGUGUAGGUUGCGUGUA .....((((.(((....((((((..((((((((((((((..((((.....))))(((((.....))))).)))))))))..))))))))))))))))))....... ( -25.90, z-score = 0.01, R) >consensus GGGUCGACACCGCGCUGGCAUCGCUCGGCAAUAAGUUGCUGAGCCUCACCAGCUACGGCCUGUUCUUGCUGU__CCAGUUAGGUAGUCUCCA__A___________ ..............((((....(((((((((....)))))))))...........((((........))))...))))............................ (-12.73 = -12.73 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:14:04 2011