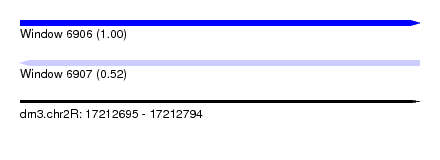
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,212,695 – 17,212,794 |
| Length | 99 |
| Max. P | 0.996471 |

| Location | 17,212,695 – 17,212,794 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 75.18 |
| Shannon entropy | 0.47233 |
| G+C content | 0.31751 |
| Mean single sequence MFE | -21.45 |
| Consensus MFE | -12.81 |
| Energy contribution | -13.06 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.94 |
| SVM RNA-class probability | 0.996471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
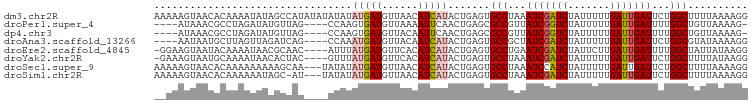
>dm3.chr2R 17212695 99 + 21146708 AAAAAGUAACACAAAAUAUAGCCAUAUAUAUAUAUGAUGUUAACAUCAUACUGAGUGCCUAAAUCGAUCUAUUUUUGAUUGAUUCUGGCUUUUAAAAGG ...............(((((......))))).(((((((....))))))).((((.(((..((((((((.......))))))))..))).))))..... ( -22.60, z-score = -3.47, R) >droPer1.super_4 2045276 90 - 7162766 ----AUAAACGCCUAGAUAUGUUAG----CCAAGUGAUGUUAAAAUUCAACUGAGCGCCGUUAUCGGUCUAUUUUUGAUUGAUUUUGGCUGUUAAAAG- ----...(((..............(----(((..(((.........)))..)).))((((..(((((((.......)))))))..)))).))).....- ( -19.50, z-score = -1.47, R) >dp4.chr3 9978006 90 - 19779522 ----AUAAACGCCUAGAUAUGUUAG----CCAAGUGAUGUUACAAUUCAACUGAGCGCCGUUAUCGGUCUAUUUUUGAUUGAUUUUGGCUGUUAAAAG- ----...(((..............(----(((..(((.........)))..)).))((((..(((((((.......)))))))..)))).))).....- ( -19.50, z-score = -1.21, R) >droAna3.scaffold_13266 5014908 91 + 19884421 ----AAUAAUGCUUAGUUAGAUCAG----CCAAAUGAUGUUACAAUCAUACUGAGUGCCGCUAUCGAUCUAUUUUUGAUUGAUUCUGGCGUAUAAAAGG ----......(((((((........----....(((((......))))))))))))((((..(((((((.......)))))))..)))).......... ( -22.20, z-score = -2.07, R) >droEre2.scaffold_4845 11356497 94 + 22589142 -GGAAGUAAUACAAAAUAACGCAAC----AUUUAUGAUGUUCACAUCAUACUGAGUGCCUGAAUCGAUCUAUUCUUGAUUGAUUUUGGCUAUUAUAAGG -....((((((..............----...(((((((....)))))))......(((.(((((((((.......))))))))).))))))))).... ( -23.40, z-score = -3.09, R) >droYak2.chr2R 9163604 94 - 21139217 -GAAAGUAAUGCAAAAUAACACUAC----GUUUAUGAUGUUCACAUCAUACUGAGUGCCUAAAUCGAUCUAUUUUUGAUUGAUUCUGGCUUUUAUAAGG -....(((.((........)).)))----...(((((((....))))))).((((.(((..((((((((.......))))))))..))).))))..... ( -22.60, z-score = -3.16, R) >droSec1.super_9 544064 96 + 3197100 AAAAAGUAACACAAAAAAAAAGCAA---UAUAUAUGAUGUUAACAUCAUACUGAGUGCCUAAAUCCAUCUAUUUUUGAUUGAUUCUGGCUUUUAAAAGG .........................---....(((((((....))))))).((((.(((..((((.(((.......))).))))..))).))))..... ( -17.80, z-score = -2.24, R) >droSim1.chr2R 15864910 95 + 19596830 AAAAAGUAACACAAAAAAUAGC-AU---UAUAUAUGAUGUUAACAUCAUACUGAGUGCCUAAAUCGAUCUAUUUUUGAUUGAUUCUGGCUUUUAAAAGG .((((((.............((-((---(...(((((((....)))))))...)))))(..((((((((.......))))))))..)))))))...... ( -24.00, z-score = -4.36, R) >consensus __AAAGUAACACAAAAAAAAGCCAG____AUAUAUGAUGUUAACAUCAUACUGAGUGCCUAAAUCGAUCUAUUUUUGAUUGAUUCUGGCUUUUAAAAGG .................................(((((......))))).......(((...(((((((.......)))))))...))).......... (-12.81 = -13.06 + 0.25)
| Location | 17,212,695 – 17,212,794 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 75.18 |
| Shannon entropy | 0.47233 |
| G+C content | 0.31751 |
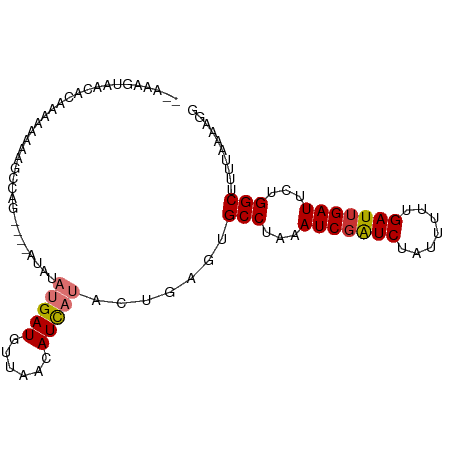
| Mean single sequence MFE | -18.29 |
| Consensus MFE | -6.93 |
| Energy contribution | -7.92 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.521197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17212695 99 - 21146708 CCUUUUAAAAGCCAGAAUCAAUCAAAAAUAGAUCGAUUUAGGCACUCAGUAUGAUGUUAACAUCAUAUAUAUAUAUGGCUAUAUUUUGUGUUACUUUUU .........((((((((((.(((.......))).))))).........((((((((....)))))))).......)))))................... ( -19.90, z-score = -2.27, R) >droPer1.super_4 2045276 90 + 7162766 -CUUUUAACAGCCAAAAUCAAUCAAAAAUAGACCGAUAACGGCGCUCAGUUGAAUUUUAACAUCACUUGG----CUAACAUAUCUAGGCGUUUAU---- -.........(((.................(.(((....))))(((.(((.((.........))))).))----)...........)))......---- ( -12.80, z-score = -0.65, R) >dp4.chr3 9978006 90 + 19779522 -CUUUUAACAGCCAAAAUCAAUCAAAAAUAGACCGAUAACGGCGCUCAGUUGAAUUGUAACAUCACUUGG----CUAACAUAUCUAGGCGUUUAU---- -.........(((.................(.(((....))))(((.(((.((.........))))).))----)...........)))......---- ( -12.80, z-score = -0.12, R) >droAna3.scaffold_13266 5014908 91 - 19884421 CCUUUUAUACGCCAGAAUCAAUCAAAAAUAGAUCGAUAGCGGCACUCAGUAUGAUUGUAACAUCAUUUGG----CUGAUCUAACUAAGCAUUAUU---- ..........(((...(((.(((.......))).)))...)))..((((((((((......)))))...)----)))).................---- ( -14.00, z-score = -0.20, R) >droEre2.scaffold_4845 11356497 94 - 22589142 CCUUAUAAUAGCCAAAAUCAAUCAAGAAUAGAUCGAUUCAGGCACUCAGUAUGAUGUGAACAUCAUAAAU----GUUGCGUUAUUUUGUAUUACUUCC- ..........(((..((((.(((.......))).))))..)))......(((((((....)))))))...----..((((......))))........- ( -18.70, z-score = -1.86, R) >droYak2.chr2R 9163604 94 + 21139217 CCUUAUAAAAGCCAGAAUCAAUCAAAAAUAGAUCGAUUUAGGCACUCAGUAUGAUGUGAACAUCAUAAAC----GUAGUGUUAUUUUGCAUUACUUUC- ..........(((.(((((.(((.......))).))))).)))......(((((((....)))))))...----(((((((......)))))))....- ( -26.60, z-score = -5.08, R) >droSec1.super_9 544064 96 - 3197100 CCUUUUAAAAGCCAGAAUCAAUCAAAAAUAGAUGGAUUUAGGCACUCAGUAUGAUGUUAACAUCAUAUAUA---UUGCUUUUUUUUUGUGUUACUUUUU ..........(((.(((((.(((.......))).))))).))).....((((((((....))))))))...---......................... ( -19.50, z-score = -2.14, R) >droSim1.chr2R 15864910 95 - 19596830 CCUUUUAAAAGCCAGAAUCAAUCAAAAAUAGAUCGAUUUAGGCACUCAGUAUGAUGUUAACAUCAUAUAUA---AU-GCUAUUUUUUGUGUUACUUUUU ..........(((.(((((.(((.......))).))))).))).....((((((((....)))))))).((---((-((........))))))...... ( -22.00, z-score = -3.66, R) >consensus CCUUUUAAAAGCCAGAAUCAAUCAAAAAUAGAUCGAUUUAGGCACUCAGUAUGAUGUUAACAUCAUAUAU____CUAGCUUUAUUUUGCGUUACUUU__ ..........(((...(((.(((.......))).)))...))).......(((((......)))))................................. ( -6.93 = -7.92 + 1.00)

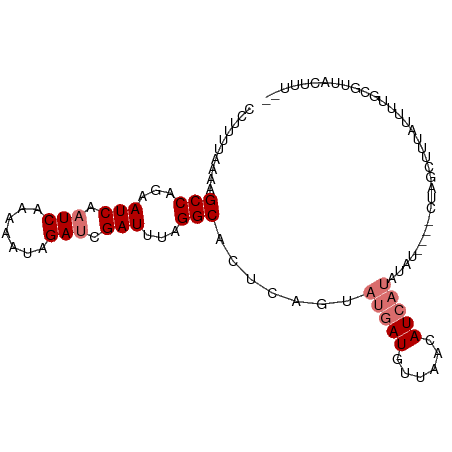
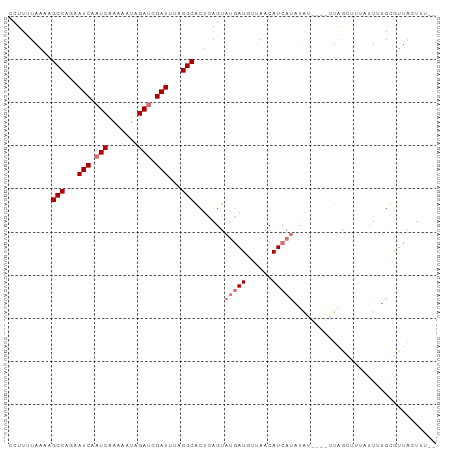
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:33 2011