| Sequence ID | dm3.chr2R |
|---|---|
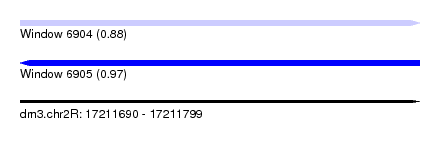
| Location | 17,211,690 – 17,211,799 |
| Length | 109 |
| Max. P | 0.972967 |

| Location | 17,211,690 – 17,211,799 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 87.63 |
| Shannon entropy | 0.28886 |
| G+C content | 0.53326 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -30.47 |
| Energy contribution | -30.86 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.883213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
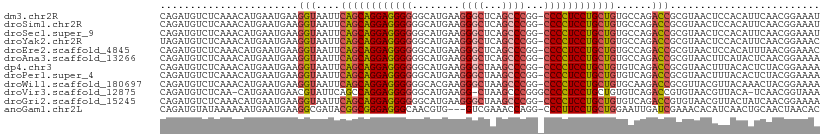
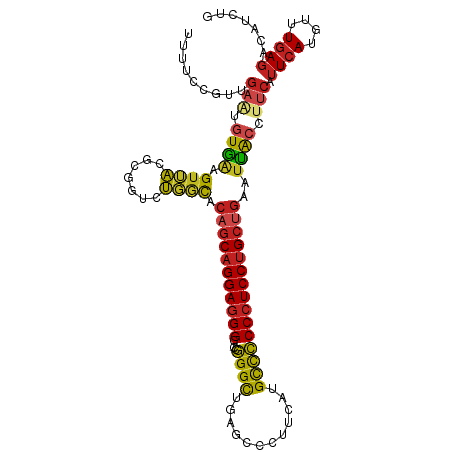
>dm3.chr2R 17211690 109 + 21146708 CAGAUGUCUCAAACAUGAAUGAAGGUAAUUCAGCAGGAGGGGGGCAUGAAGGGCUCAGCCCGG-CCCCUCCUGCUGUGCCAGACCGCGUAACUCCACAUUCAACGGAAAU ...............((((((..((((...(((((((((((((((.(((.....))))))...-)))))))))))))))).(....).........))))))........ ( -39.50, z-score = -1.63, R) >droSim1.chr2R 15863899 109 + 19596830 CAGAUGUCUCAAACAUGAAUGAAGGUAAUUCAGCAGGAGGGGGGCAUGAAGGGCUCAGCCCGG-CCCCUCCUGCUGUGCCAGACCGCGUAACUCCACAUUCAACGGAAAU ...............((((((..((((...(((((((((((((((.(((.....))))))...-)))))))))))))))).(....).........))))))........ ( -39.50, z-score = -1.63, R) >droSec1.super_9 543050 109 + 3197100 CAGAUGUCUCAAACAUGAAUGAAGGUAAUUCAGCAGGAGGGGGGCAUGAAGGGCUCAGCCCGG-CCCCUCCUGCUGUGCCAGACCGCGUAACUCCACAUUCAACGGAAAU ...............((((((..((((...(((((((((((((((.(((.....))))))...-)))))))))))))))).(....).........))))))........ ( -39.50, z-score = -1.63, R) >droYak2.chr2R 9162584 109 - 21139217 UAGAUGUCUCAAACAUGAAUGAAGGUAAUUCAGCAGGAGGGGGGCAUGAAGGGCUCAGCCCGG-CCCCUCCUGCUGUGCCAGACCGCGUAACUCCACAUUCAACGGAAAC ...............((((((..((((...(((((((((((((((.(((.....))))))...-)))))))))))))))).(....).........))))))..(....) ( -39.60, z-score = -1.87, R) >droEre2.scaffold_4845 11355475 109 + 22589142 CAGAUGUCUCAAACAUGAAUGAAGGUAAUUCAGCAGGAGGGGGGCAUGAAGGGCUCAGCCCGG-CCCCUCCUGCUGUGCCAGACCGCGUAACUCCACAUUUAACGGAAAC ..(..((((((....))).....((((...(((((((((((((((.(((.....))))))...-)))))))))))))))).)))..).....(((.........)))... ( -38.80, z-score = -1.67, R) >droAna3.scaffold_13266 5013863 109 + 19884421 CAGAUGUCUCAAACAUGAAUGAAGGUAAUUCAGCAGGAGGGGGGCAUGAAGGGCUCAGCCCGG-CCCCUCCUGCUGUGCCAGACCGCGUAACUUCAUACUCAACGGAAAA ...............(((((((((......(((((((((((((((.(((.....))))))...-)))))))))))).((......))....))))))..)))........ ( -39.10, z-score = -1.66, R) >dp4.chr3 9976957 109 - 19779522 CAGAUGUCUCAAACAUGAAUGAAGGUAAUUCAGCAGGAGGGGGGCAUGAAGGGCUAAGCCCGG-CCCCUCCUGCUGUGUCAGACCGCGUAACUUUACACUCUACGGAAAA ...((((.(((....))).....(((....((((((((..(((((.....((((...)))).)-))))))))))))......)))))))..........((....))... ( -37.30, z-score = -1.53, R) >droPer1.super_4 2044218 109 - 7162766 CAGAUGUCUCAAACAUGAAUGAAGGUAAUUCAGCAGGAGGGGGGCAUGAAGGGCUAAGCCCGG-CCCCUCCUGCUGUGUCAGACCGCGUAACUUUACACUCUACGGAAAA ...((((.(((....))).....(((....((((((((..(((((.....((((...)))).)-))))))))))))......)))))))..........((....))... ( -37.30, z-score = -1.53, R) >droWil1.scaffold_180697 1465779 109 + 4168966 CAGAUGUCUCAAACAUGAAUGAAGGUAAUUCAGCAGGAGGGGGGCACGAAGGGCUAAGCCCGG-CCCCUCCUGCUGUGCAAGACCGCGUUACGUUACAAACUACGGAAAA ..(((((.(((....))).....(((....((((((((..(((((.....((((...)))).)-))))))))))))......)))))))).(((........)))..... ( -38.60, z-score = -2.06, R) >droVir3.scaffold_12875 2542493 107 - 20611582 CAGAUGUCUCAA-CAUGAAUGAACGUAUUCAGCCAGGAGGGGGGCAUGAAGG-CUAAGCCCGGGCCCCUCCUGCUGUGUCAGACCGUGUAACGUUACA-UCAACGGUAAA ..(((((..(.(-((((..(((.......((((.(((((((((((.......-....)))....))))))))))))..)))...)))))...)..)))-))......... ( -35.10, z-score = -0.91, R) >droGri2.scaffold_15245 14127781 109 - 18325388 CAGAUGUCUCAAACAUGAAUGAAGGUAAUUCAGCAGGAGGGGGGCAUGAAGGGCUAAGCCCGG-CCCCUCCUGCUGUGUCAGACCGUGUAACGUUACUAUCAACGGAAAA ............(((((..(((........((((((((..(((((.....((((...)))).)-))))))))))))..)))...)))))..((((......))))..... ( -38.00, z-score = -1.91, R) >anoGam1.chr2L 26876184 106 + 48795086 CAGAUGUAUAAAAAAUGAAUGAAGGCGAUACGGCGGGAGGGCAACGUG---GUCGAAACCAGG-CCCUUCCUGCUGGAAUUGAUCGAAACACAUCAACUGCAACUAACAC ..(((((..................((((.((((((((((((....((---((....)))).)-))).))))))))......))))....)))))............... ( -30.15, z-score = -2.40, R) >consensus CAGAUGUCUCAAACAUGAAUGAAGGUAAUUCAGCAGGAGGGGGGCAUGAAGGGCUAAGCCCGG_CCCCUCCUGCUGUGCCAGACCGCGUAACUUCACAUUCAACGGAAAA .......................(((....((((((((((((........((((...))))...))))))))))))......)))......................... (-30.47 = -30.86 + 0.38)

| Location | 17,211,690 – 17,211,799 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 87.63 |
| Shannon entropy | 0.28886 |
| G+C content | 0.53326 |
| Mean single sequence MFE | -39.11 |
| Consensus MFE | -29.38 |
| Energy contribution | -29.29 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.972967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
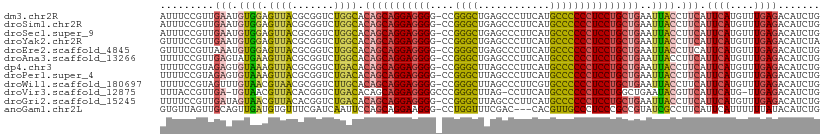
>dm3.chr2R 17211690 109 - 21146708 AUUUCCGUUGAAUGUGGAGUUACGCGGUCUGGCACAGCAGGAGGGG-CCGGGCUGAGCCCUUCAUGCCCCCCUCCUGCUGAAUUACCUUCAUUCAUGUUUGAGACAUCUG .((((...((((((.((((((..((......)).((((((((((((-...(((((((...)))).))))))))))))))))))).))..)))))).....))))...... ( -42.90, z-score = -2.57, R) >droSim1.chr2R 15863899 109 - 19596830 AUUUCCGUUGAAUGUGGAGUUACGCGGUCUGGCACAGCAGGAGGGG-CCGGGCUGAGCCCUUCAUGCCCCCCUCCUGCUGAAUUACCUUCAUUCAUGUUUGAGACAUCUG .((((...((((((.((((((..((......)).((((((((((((-...(((((((...)))).))))))))))))))))))).))..)))))).....))))...... ( -42.90, z-score = -2.57, R) >droSec1.super_9 543050 109 - 3197100 AUUUCCGUUGAAUGUGGAGUUACGCGGUCUGGCACAGCAGGAGGGG-CCGGGCUGAGCCCUUCAUGCCCCCCUCCUGCUGAAUUACCUUCAUUCAUGUUUGAGACAUCUG .((((...((((((.((((((..((......)).((((((((((((-...(((((((...)))).))))))))))))))))))).))..)))))).....))))...... ( -42.90, z-score = -2.57, R) >droYak2.chr2R 9162584 109 + 21139217 GUUUCCGUUGAAUGUGGAGUUACGCGGUCUGGCACAGCAGGAGGGG-CCGGGCUGAGCCCUUCAUGCCCCCCUCCUGCUGAAUUACCUUCAUUCAUGUUUGAGACAUCUA (((((...((((((.((((((..((......)).((((((((((((-...(((((((...)))).))))))))))))))))))).))..)))))).....)))))..... ( -46.50, z-score = -3.44, R) >droEre2.scaffold_4845 11355475 109 - 22589142 GUUUCCGUUAAAUGUGGAGUUACGCGGUCUGGCACAGCAGGAGGGG-CCGGGCUGAGCCCUUCAUGCCCCCCUCCUGCUGAAUUACCUUCAUUCAUGUUUGAGACAUCUG (((((........((((((....((......)).((((((((((((-...(((((((...)))).)))))))))))))))......))))))........)))))..... ( -43.29, z-score = -2.82, R) >droAna3.scaffold_13266 5013863 109 - 19884421 UUUUCCGUUGAGUAUGAAGUUACGCGGUCUGGCACAGCAGGAGGGG-CCGGGCUGAGCCCUUCAUGCCCCCCUCCUGCUGAAUUACCUUCAUUCAUGUUUGAGACAUCUG .((((...(((..((((((....((......)).((((((((((((-...(((((((...)))).)))))))))))))))......))))))))).....))))...... ( -40.20, z-score = -2.25, R) >dp4.chr3 9976957 109 + 19779522 UUUUCCGUAGAGUGUAAAGUUACGCGGUCUGACACAGCAGGAGGGG-CCGGGCUUAGCCCUUCAUGCCCCCCUCCUGCUGAAUUACCUUCAUUCAUGUUUGAGACAUCUG .......(((((((((....))))).((((.(.(((((((((((((-..((((...))))........))))))))))(((((.......)))))))).).)))).)))) ( -39.00, z-score = -2.73, R) >droPer1.super_4 2044218 109 + 7162766 UUUUCCGUAGAGUGUAAAGUUACGCGGUCUGACACAGCAGGAGGGG-CCGGGCUUAGCCCUUCAUGCCCCCCUCCUGCUGAAUUACCUUCAUUCAUGUUUGAGACAUCUG .......(((((((((....))))).((((.(.(((((((((((((-..((((...))))........))))))))))(((((.......)))))))).).)))).)))) ( -39.00, z-score = -2.73, R) >droWil1.scaffold_180697 1465779 109 - 4168966 UUUUCCGUAGUUUGUAACGUAACGCGGUCUUGCACAGCAGGAGGGG-CCGGGCUUAGCCCUUCGUGCCCCCCUCCUGCUGAAUUACCUUCAUUCAUGUUUGAGACAUCUG .....(((.(((........))))))((((((.(((((((((((((-..((((...))))........))))))))))(((((.......)))))))).))))))..... ( -36.80, z-score = -2.22, R) >droVir3.scaffold_12875 2542493 107 + 20611582 UUUACCGUUGA-UGUAACGUUACACGGUCUGACACAGCAGGAGGGGCCCGGGCUUAG-CCUUCAUGCCCCCCUCCUGGCUGAAUACGUUCAUUCAUG-UUGAGACAUCUG ((((.(((.((-((.(((((.....(......).((((((((((((....(((....-.......))))))))))).))))...))))))))).)))-.))))....... ( -34.00, z-score = -1.21, R) >droGri2.scaffold_15245 14127781 109 + 18325388 UUUUCCGUUGAUAGUAACGUUACACGGUCUGACACAGCAGGAGGGG-CCGGGCUUAGCCCUUCAUGCCCCCCUCCUGCUGAAUUACCUUCAUUCAUGUUUGAGACAUCUG ........(((..((((.((((.......)))).((((((((((((-..((((...))))........))))))))))))..))))..)))((((....))))....... ( -36.00, z-score = -2.55, R) >anoGam1.chr2L 26876184 106 - 48795086 GUGUUAGUUGCAGUUGAUGUGUUUCGAUCAAUUCCAGCAGGAAGGG-CCUGGUUUCGAC---CACGUUGCCCUCCCGCCGUAUCGCCUUCAUUCAUUUUUUAUACAUCUG ((((..((((.(((((((........))))))).)))).((.((((-(.((((....))---))....))))).)).........................))))..... ( -25.80, z-score = -1.25, R) >consensus UUUUCCGUUGAAUGUGAAGUUACGCGGUCUGGCACAGCAGGAGGGG_CCGGGCUGAGCCCUUCAUGCCCCCCUCCUGCUGAAUUACCUUCAUUCAUGUUUGAGACAUCUG .........(((.((((.((((.......)))).(((((((((((....((((............)))))))))))))))..)))).))).((((....))))....... (-29.38 = -29.29 + -0.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:31 2011