| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,200,084 – 17,200,176 |
| Length | 92 |
| Max. P | 0.545100 |

| Location | 17,200,084 – 17,200,176 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 58.75 |
| Shannon entropy | 0.74516 |
| G+C content | 0.44132 |
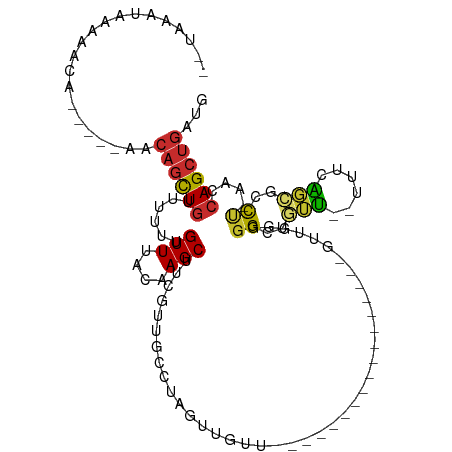
| Mean single sequence MFE | -24.70 |
| Consensus MFE | -6.99 |
| Energy contribution | -6.48 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.545100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
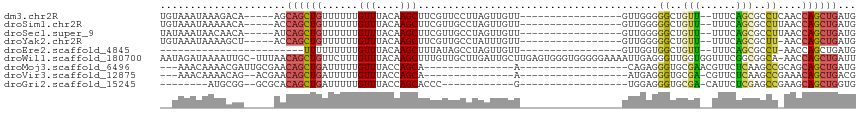
>dm3.chr2R 17200084 92 + 21146708 UGUAAAUAAAGACA-----AGCAGCUGUUUUUUGUUUACAAGCUUCGUUCCUUAGUUGUU-----------------GUUGGGGGCUGUU--UUUCAGCGCCUCAACCAGCUGAUG ((((((((((((((-----......)).))))))))))))...........(((((((..-----------------(((((((((((..--...)))).)))))))))))))).. ( -31.50, z-score = -3.30, R) >droSim1.chr2R 15851988 92 + 19596830 UGUAAAUAAAAACA-----ACCAGCUGUUUUUUGUUUACAAGCUUCGUUGCCUAGUUGUU-----------------GUUGGGGGCUGUU--UUUCAGCGCCUUAACCAGCUGAUG ((((((((((((((-----......)).)))))))))))).((......)).((((((..-----------------(((((((((((..--...)))).)))))))))))))... ( -29.20, z-score = -2.66, R) >droSec1.super_9 529864 92 + 3197100 UAUAAAUAACAACA-----AUCAGCUGUUUUUUGUUUACAAGCUUCGUUGCCUAGUUGUU-----------------GUUGGGGGCUGUU--UUUCAGCGCCUUAACCAGCUGAUG ..............-----((((((((..............((......)).........-----------------(((((((((((..--...)))).))))))))))))))). ( -26.90, z-score = -2.34, R) >droYak2.chr2R 9150940 91 - 21139217 UGUAAAUAAAAGCU-----ACCAGCUGUUUUUUGUUUACAAGCUUCGUUGCCUAUUUGUU-----------------GUUGGGGGCUGUU--UUUCAGCGCUU-AACCAGCUGAUG (((((((((((((.-----....)))....)))))))))).((......)).((((.(((-----------------((((((.((((..--...)))).)))-)).)))).)))) ( -25.10, z-score = -1.44, R) >droEre2.scaffold_4845 11343801 72 + 22589142 ------------------------UUUUUUUUUGUUUACAAGCUUUAUAGCCUAGUUGUU-----------------GUUGGUGGCUGUU--UUUCAGCGCCU-AACCAGCUGAUG ------------------------.................(((....))).((((((..-----------------(((((..((((..--...))))..))-)))))))))... ( -16.80, z-score = -1.29, R) >droWil1.scaffold_180700 5684422 114 - 6630534 AAUAGAUAAAAUUGC-UUUAACAGCUGUUCUUUGUUUACAAGCUUUGUUGCUUGAUUGCUUGAGUGGGUGGGGGAAAAUUGAGGGUUGGUGGUUUCGGCGGCA-AACCAGCUGAUU ...............-.....(((((.(((((..(((((((((...(((....))).))))..)))))..)))))........((((.((.((....)).)).-)))))))))... ( -26.70, z-score = 0.41, R) >droMoj3.scaffold_6496 14157247 80 - 26866924 ---AAACAAAACGAUUGCGAACAGCUGAUUUUUGUUUACCAGCA---------------A------------------CAGAGGGUGCGAACGUUCUCAAGCCGCAGCAGCUGAUG ---((((((((.....((.....))....))))))))..((((.---------------.------------------..(((..((....))..)))..((....)).))))... ( -19.50, z-score = -0.24, R) >droVir3.scaffold_12875 1585291 77 - 20611582 ---AAACAAAACAG--ACGAACAGCUGAUUUUUGUUUACCAGCA---------------A------------------AUGAGGGUGCGA-CGUUCUCAAGCCGAAACAGCUGACG ---...........--.....((((((...(((((......)))---------------)------------------)((((..((...-))..))))........))))))... ( -18.30, z-score = -1.21, R) >droGri2.scaffold_15245 13237460 75 - 18325388 --------AUGCGG--GCGCACAGCUGAUUUUUGUUUACCAGCACCC------------G------------------UGGAGGGUGCGA-CAUUCUCGAGCCGAAGCAGCUGGUG --------.(((..--..)))((((((.((((.((((....((((((------------.------------------....))))))((-.....)))))).))))))))))... ( -28.30, z-score = -1.36, R) >consensus __UAAAUAAAAACA_____AACAGCUGUUUUUUGUUUACAAGCUUCGUUGCCUAGUUGUU_________________GUUGGGGGCUGUU__UUUCAGCGCCU_AACCAGCUGAUG .....................((((((......(((....)))........................................((..(((......)))..))....))))))... ( -6.99 = -6.48 + -0.51)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:29 2011