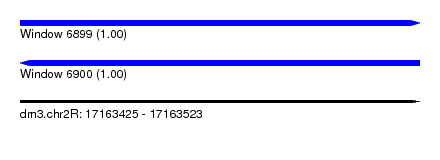
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,163,425 – 17,163,523 |
| Length | 98 |
| Max. P | 0.998391 |

| Location | 17,163,425 – 17,163,523 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 63.64 |
| Shannon entropy | 0.70365 |
| G+C content | 0.59425 |
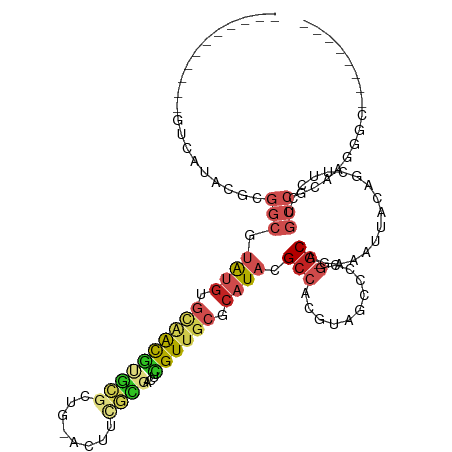
| Mean single sequence MFE | -50.20 |
| Consensus MFE | -8.60 |
| Energy contribution | -8.67 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.50 |
| Mean z-score | -4.31 |
| Structure conservation index | 0.17 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.998391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
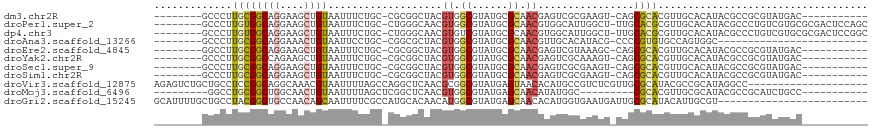
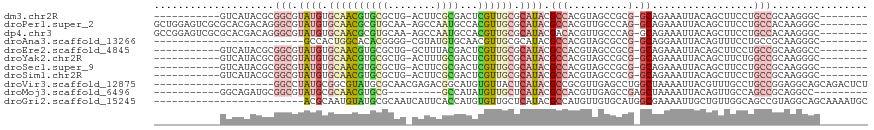
>dm3.chr2R 17163425 98 + 21146708 --------GCCCUUGCGGCAGGAAGCUGUAAUUUCUGC-CGCGGCUACGUGGCGUAUGCGCAACGAGUCGCGAAGU-CAGCGCACGUUGCACAUACGCCGCGUAUGAC----------- --------(((...((((((((((.......)))))))-))))))(((((((((((((.((((((.(.(((.....-..)))).)))))).)))))))))))))....----------- ( -58.80, z-score = -6.36, R) >droPer1.super_2 3820013 109 + 9036312 --------GCCCUUGUGGCAGGAAGCUGUAAUUUCUGC-CUGGGCAACGUGGCGUAUGCGCAACGUGGCAUUGGCU-UUGCACGCGUUGCACAUACGCCCUGUCGUGCGCGACUCCAGC --------((((....((((((((.......)))))))-).))))...(.((((((((.(((((((((((......-.))).)))))))).))))))))).((((....))))...... ( -56.70, z-score = -4.06, R) >dp4.chr3 3629765 109 + 19779522 --------GCCCUUGUGGCAGGAAGCUGUAAUUUCUGC-CUGGGCAACGUGUCGUAUGCGCAACGUGGCAUUGGCU-UUGCACGCGUUGCACAUACGCCCUGUCGUGCGCGACUCCGGC --------((((....((((((((.......)))))))-).))))........(((((.(((((((((((......-.))).)))))))).)))))(((..((((....))))...))) ( -52.80, z-score = -3.20, R) >droAna3.scaffold_13266 4961980 84 + 19884421 --------GCCCUUGCGGCAGGAAACUGUAAUUCCUGC-CGGCGCUACGUGGCGUAUGCGCAACGUUGCACAUACG-CCCCGUGUGCCAGUGGC------------------------- --------(((((...((((((((.......)))))))-)(((((.(((.((((((((.(((....))).))))))-)).)))))))))).)))------------------------- ( -48.90, z-score = -5.26, R) >droEre2.scaffold_4845 11307512 98 + 22589142 --------GGCCUUGCGGCAGGAAGCUGUAAUUUCUGC-CGCGGCUACGUGGCGUAUGCGCAACGAGUCGUAAAGC-CAGCGCACGUUGCACAUACGCCGCGUAUGAC----------- --------((((..((((((((((.......)))))))-)))))))((((((((((((.((((((.(.(((.....-..)))).)))))).)))))))))))).....----------- ( -56.60, z-score = -5.79, R) >droYak2.chr2R 9114291 98 - 21139217 --------GCCCUUGCGGCCAGAAGCUGUAAUUUCUGC-CGCGGCUACGUGGCGUAUGCGCAACGAGUCGCAAAGU-CAGCGCACGUUGCACAUACGCCGCGUAUGAC----------- --------(((...(((((.(((((......)))))))-))))))(((((((((((((.((((((.(.(((.....-..)))).)))))).)))))))))))))....----------- ( -54.00, z-score = -5.55, R) >droSec1.super_9 493389 98 + 3197100 --------GCCCUUGCGGCAGGAAGCUGUAAUUUCUGC-CGCGGCUACGUGGCGUAUGCGCAACGAGUCGCGAAGU-CAGCGCACGUUGCACAUACGCCGCGUAUGAC----------- --------(((...((((((((((.......)))))))-))))))(((((((((((((.((((((.(.(((.....-..)))).)))))).)))))))))))))....----------- ( -58.80, z-score = -6.36, R) >droSim1.chr2R 15814922 98 + 19596830 --------GCCCUUGCGGCAGGAAGCUGUAAUUUCUGC-CGCGGCUACGUGGCGUAUGCGCAACGAGUCGCGAAGU-CAGCGCACGUUGCACAUACGCCGCGUAUGAC----------- --------(((...((((((((((.......)))))))-))))))(((((((((((((.((((((.(.(((.....-..)))).)))))).)))))))))))))....----------- ( -58.80, z-score = -6.36, R) >droVir3.scaffold_12875 14347912 99 + 20611582 AGAGUCUGCUGCCUCCGGCAGGCAAACGUAAUUUUAGCCAGGCUCAACGCGGCGUAUGAGUAACACAUGCCGUCUCGUUGCGCAUACGCCGCAUAGGCC-------------------- .((((((((((((...)))))(((((......))).)))))))))...((((((((((.(((((............))))).)))))))))).......-------------------- ( -41.60, z-score = -2.97, R) >droMoj3.scaffold_6496 9809200 90 + 26866924 ---------GGCCUGCGGCUGGCAACUGUAAUUUUAGCUCGGCUCAACGUGGCGUAUGAGCAACAUAUGGC---------CGCACGUUGCGCAUACGCCGCAUCUGCC----------- ---------(((.((.(((((((.............)).)))))))..((((((((((.(((((.......---------.....))))).))))))))))....)))----------- ( -34.92, z-score = -0.82, R) >droGri2.scaffold_15245 14331355 94 + 18325388 GCAUUUUGCUGCCUACGGCUGCCAACAGCAAUUUUCGCCAUGCACAACAUGGCGUAUGAGCAACACAUGGUGAAUGAUUGCGCAUACAUUGCGU------------------------- .......((.(((...))).)).....(((((....((((((.....))))))(((((.((((..(((.....))).)))).))))))))))..------------------------- ( -30.30, z-score = -0.70, R) >consensus ________GCCCUUGCGGCAGGAAGCUGUAAUUUCUGC_CGGGGCUACGUGGCGUAUGCGCAACGAGUCGCGAAGU_CAGCGCACGUUGCACAUACGCCGCGUAUGAC___________ .............((((((((....))))...................((.((......)).))................))))................................... ( -8.60 = -8.67 + 0.08)
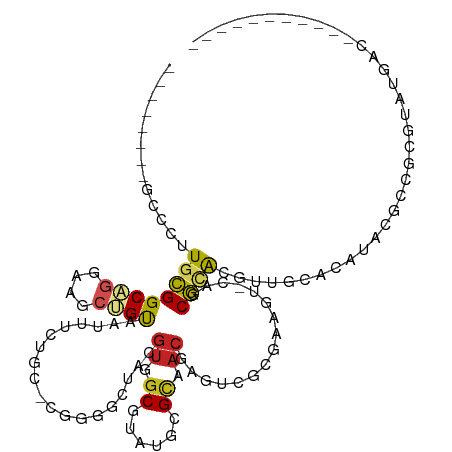
| Location | 17,163,425 – 17,163,523 |
|---|---|
| Length | 98 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 63.64 |
| Shannon entropy | 0.70365 |
| G+C content | 0.59425 |
| Mean single sequence MFE | -47.83 |
| Consensus MFE | -8.70 |
| Energy contribution | -9.68 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.68 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.18 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.995994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17163425 98 - 21146708 -----------GUCAUACGCGGCGUAUGUGCAACGUGCGCUG-ACUUCGCGACUCGUUGCGCAUACGCCACGUAGCCGCG-GCAGAAAUUACAGCUUCCUGCCGCAAGGGC-------- -----------....((((.(((((((((((((((.((((..-.....))).).))))))))))))))).))))((((((-((((.............)))))))...)))-------- ( -53.92, z-score = -5.33, R) >droPer1.super_2 3820013 109 - 9036312 GCUGGAGUCGCGCACGACAGGGCGUAUGUGCAACGCGUGCAA-AGCCAAUGCCACGUUGCGCAUACGCCACGUUGCCCAG-GCAGAAAUUACAGCUUCCUGCCACAAGGGC-------- ((((..((((....))))..(((((((((((((((.(.(((.-......)))).)))))))))))))))...((((....-))))......)))).((((......)))).-------- ( -49.20, z-score = -2.81, R) >dp4.chr3 3629765 109 - 19779522 GCCGGAGUCGCGCACGACAGGGCGUAUGUGCAACGCGUGCAA-AGCCAAUGCCACGUUGCGCAUACGACACGUUGCCCAG-GCAGAAAUUACAGCUUCCUGCCACAAGGGC-------- (((...((((....)))).((((((((((((((((.(.(((.-......)))).))))))))))))((....)))))).(-((((.............))))).....)))-------- ( -45.42, z-score = -2.15, R) >droAna3.scaffold_13266 4961980 84 - 19884421 -------------------------GCCACUGGCACACGGGG-CGUAUGUGCAACGUUGCGCAUACGCCACGUAGCGCCG-GCAGGAAUUACAGUUUCCUGCCGCAAGGGC-------- -------------------------(((.(((((.((((.((-((((((((((....)))))))))))).))).).)))(-(((((((.......))))))))...)))))-------- ( -50.10, z-score = -5.89, R) >droEre2.scaffold_4845 11307512 98 - 22589142 -----------GUCAUACGCGGCGUAUGUGCAACGUGCGCUG-GCUUUACGACUCGUUGCGCAUACGCCACGUAGCCGCG-GCAGAAAUUACAGCUUCCUGCCGCAAGGCC-------- -----------....((((.(((((((((((((((.(((...-......)).).))))))))))))))).))))((((((-((((.............)))))))..))).-------- ( -50.92, z-score = -4.75, R) >droYak2.chr2R 9114291 98 + 21139217 -----------GUCAUACGCGGCGUAUGUGCAACGUGCGCUG-ACUUUGCGACUCGUUGCGCAUACGCCACGUAGCCGCG-GCAGAAAUUACAGCUUCUGGCCGCAAGGGC-------- -----------....((((.(((((((((((((((.((((..-.....))).).))))))))))))))).))))((((((-((((((........)))).)))))...)))-------- ( -54.20, z-score = -5.29, R) >droSec1.super_9 493389 98 - 3197100 -----------GUCAUACGCGGCGUAUGUGCAACGUGCGCUG-ACUUCGCGACUCGUUGCGCAUACGCCACGUAGCCGCG-GCAGAAAUUACAGCUUCCUGCCGCAAGGGC-------- -----------....((((.(((((((((((((((.((((..-.....))).).))))))))))))))).))))((((((-((((.............)))))))...)))-------- ( -53.92, z-score = -5.33, R) >droSim1.chr2R 15814922 98 - 19596830 -----------GUCAUACGCGGCGUAUGUGCAACGUGCGCUG-ACUUCGCGACUCGUUGCGCAUACGCCACGUAGCCGCG-GCAGAAAUUACAGCUUCCUGCCGCAAGGGC-------- -----------....((((.(((((((((((((((.((((..-.....))).).))))))))))))))).))))((((((-((((.............)))))))...)))-------- ( -53.92, z-score = -5.33, R) >droVir3.scaffold_12875 14347912 99 - 20611582 --------------------GGCCUAUGCGGCGUAUGCGCAACGAGACGGCAUGUGUUACUCAUACGCCGCGUUGAGCCUGGCUAAAAUUACGUUUGCCUGCCGGAGGCAGCAGACUCU --------------------((((.((((((((((((.(.((((..........)))).).)))))))))))).).))).............((((((.((((...))))))))))... ( -40.40, z-score = -1.90, R) >droMoj3.scaffold_6496 9809200 90 - 26866924 -----------GGCAGAUGCGGCGUAUGCGCAACGUGCG---------GCCAUAUGUUGCUCAUACGCCACGUUGAGCCGAGCUAAAAUUACAGUUGCCAGCCGCAGGCC--------- -----------(((...(((((((((((.((((((((..---------....)))))))).))))))))..((((.((..((((........)))))))))).))).)))--------- ( -39.10, z-score = -2.33, R) >droGri2.scaffold_15245 14331355 94 - 18325388 -------------------------ACGCAAUGUAUGCGCAAUCAUUCACCAUGUGUUGCUCAUACGCCAUGUUGUGCAUGGCGAAAAUUGCUGUUGGCAGCCGUAGGCAGCAAAAUGC -------------------------..(((..(((((.((((((((.....))).))))).)))))((((((.....)))))).....(((((((..((....))..)))))))..))) ( -35.00, z-score = -1.33, R) >consensus ___________GUCAUACGCGGCGUAUGUGCAACGUGCGCUG_ACUUCGCGACUCGUUGCGCAUACGCCACGUAGCCCCG_GCAGAAAUUACAGCUUCCUGCCGCAAGGGC________ ....................(((.((((.((((((((((........))))...)))))).)))).........((.....)).................)))................ ( -8.70 = -9.68 + 0.98)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:27 2011