| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,142,620 – 17,142,713 |
| Length | 93 |
| Max. P | 0.966504 |

| Location | 17,142,620 – 17,142,713 |
|---|---|
| Length | 93 |
| Sequences | 8 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 79.26 |
| Shannon entropy | 0.41311 |
| G+C content | 0.39927 |
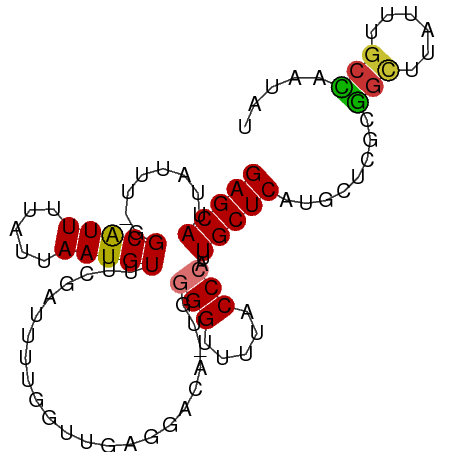
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -15.20 |
| Energy contribution | -14.83 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.33 |
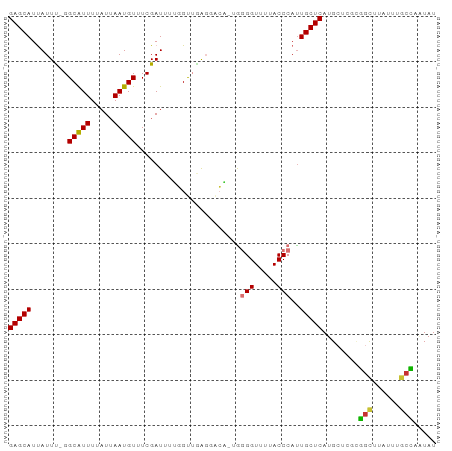
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
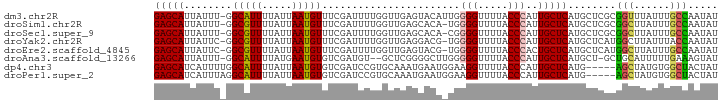
>dm3.chr2R 17142620 93 - 21146708 GAGCAUUAUUU-GGCAUUUUAUUAAUGUUUCGAUUUUGGUUGAGUACAUUGGGGUUUUACCCAUUGCUCAUGCUCGCGGUUUAUUUGCCAAUAU ((((((.....-((((......((((((((((((....)))))).))))))(((.....)))..)))).))))))..(((......)))..... ( -25.90, z-score = -2.71, R) >droSim1.chr2R 15793591 92 - 19596830 GAGCAUUAUUU-GGCGUUUUAUUAAUGUUUCGAUUUUGGUUGAGCACA-UGGGGUUUUACCCAUUGCUCAUGCUCGCGGCUUAUUUGCCAAUAU (((((......-((((((.....))))))...........((((((.(-((((......))))))))))))))))..(((......)))..... ( -30.80, z-score = -3.65, R) >droSec1.super_9 472923 92 - 3197100 GAGCAUUAUUU-GGCGUUUUAUUAAUGUUUCGAUUUUGGUUGAGCACA-CGGGGUUUUACCCAUUGCUCAUGCUCGCGGCUUAUUUGCCAAUAU (((((......-((((((.....))))))...........((((((..-.(((......)))..)))))))))))..(((......)))..... ( -27.50, z-score = -2.61, R) >droYak2.chr2R 9092749 92 + 21139217 GAGCAUUAUUC-GGCGUUUUAUUAAUGUUUCGAUUUUGGUUGAGGACG-UGGGGUUUUACCCAUUGCUCAUGCUCAUGGCUUAUUUACCAAUAU (((((....((-((((((.....)))))..))).......((((.(.(-((((......)))))).))))))))).(((........))).... ( -24.00, z-score = -1.86, R) >droEre2.scaffold_4845 11286492 92 - 22589142 GAGCAUUAUUC-GGCGUUUUAUUAAUGUUUCGAUUUUGGUUGAGUACG-UGGGGUUUUACCCACUGCUCAUGCUCAUGGCUUAUUUGCCAAUAU (((((....((-((((((.....)))))..))).......((((((.(-((((......)))))))))))))))).((((......)))).... ( -33.30, z-score = -4.93, R) >droAna3.scaffold_13266 4937672 90 - 19884421 GAGCAUUAUUU-GGCAUUUUAUGAAUGUGUCGAUGU--GCUCGGGGCUUGGGGGUUUUACCCAUUGCUCAUGCU-GCUGCAUUUUUGAAAGUAU ((((((...((-(((((.........))))))).))--)))).((((...(((......)))...))))((((.-...))))............ ( -30.00, z-score = -2.51, R) >dp4.chr3 3611135 89 - 19779522 GAGCAUCAUUUUGGCAUUUUAUUAAUGUGUCGAUCCGUGCAAAUGAAUGGAAGGUUUUACCCAUUGCUCAUG-----AGCUAUGUGGCUACUAU (((((.....(((((((.........)))))))(((((........))))).((......))..)))))...-----((((....))))..... ( -20.70, z-score = -0.36, R) >droPer1.super_2 3801349 89 - 9036312 GAGCAUCAUUUAGGCAUUUUAUUAAUGUGUCGAUCCGUGCAAAUGAAUGGAAGGUUUUACCCAUUGCUCAUG-----AGCUAUGUGGCUACUAU (((((.......(((((.........)))))..(((((........))))).((......))..)))))...-----((((....))))..... ( -19.60, z-score = -0.02, R) >consensus GAGCAUUAUUU_GGCAUUUUAUUAAUGUUUCGAUUUUGGUUGAGGACA_UGGGGUUUUACCCAUUGCUCAUGCUCGCGGCUUAUUUGCCAAUAU (((((........(((((.....))))).......................(((.....)))..)))))........(((......)))..... (-15.20 = -14.83 + -0.37)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:23 2011