| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,111,852 – 17,111,953 |
| Length | 101 |
| Max. P | 0.987356 |

| Location | 17,111,852 – 17,111,953 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 69.58 |
| Shannon entropy | 0.58661 |
| G+C content | 0.46122 |
| Mean single sequence MFE | -22.37 |
| Consensus MFE | -11.13 |
| Energy contribution | -10.52 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.987356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17111852 101 - 21146708 -CAACACCCA-ACACCCACACCCACUUCUACAUCC---------ACAUC--CACACAAAGUGUGCUUUCAAUCAGG---CACUGAAUUGAAAAUCCAUUAACGAGUACAAGGCGGAG -.........-.........((..(((.(((....---------.....--(((((...))))).((((((((((.---..))).)))))))............))).)))..)).. ( -15.70, z-score = -1.07, R) >droSim1.chr2R 15762797 94 - 19596830 ---------CCACACCCACACCCACUUCUACAUCC---------ACAUC--CACACAAAGUGUGCUUUCAAUCAGG---CACUGAAUUGAAAAUCCAUUAACGAGUACAAGGCGGAG ---------...........((..(((.(((....---------.....--(((((...))))).((((((((((.---..))).)))))))............))).)))..)).. ( -15.70, z-score = -1.12, R) >droYak2.chr2R 9061134 95 + 21139217 --------CCAACCUCCUCCUCCACCUCCAUAUCC---------ACAUC--CACACAAAGUGUGCUUUCAAUCAGG---CACUGAAUUGAAAAUCCAUUAACGAGUACAAGGCGGAG --------.....((((.(((....(((.......---------.....--(((((...))))).((((((((((.---..))).)))))))..........)))....))).)))) ( -20.90, z-score = -2.87, R) >droEre2.scaffold_4845 11255475 103 - 22589142 CAGAGGCGCCCACCUCCACCGCCACCUCCGCCUCC---------ACAUC--CACACAAAGUGUGCUUUCAAUCAGG---CACUGAAUUGAAAAUCCAUUAACGAGUACAAGGCGGAG ....((((...........))))..((((((((..---------...((--(((((...))))).((((((((((.---..))).)))))))..........)).....)))))))) ( -29.90, z-score = -3.47, R) >droAna3.scaffold_13266 4902548 113 - 19884421 -CAACACACACACCGACACACACACUCGCACAGUCUGGUGGGCAACACCCACACACAAAGUGUGCUUUCAAUCAGG---CACUGAAUUGAAAAUCCAUUAACGAGUACAAGGCAGAG -......................(((((((((.(.(((((((.....))))).))...).)))))((((((((((.---..))).)))))))..........))))........... ( -26.00, z-score = -1.49, R) >dp4.chr3 3581786 95 - 19779522 ---CUGUAAACACAGUGCCGUACAUACGUACAGUGU--------ACGC---CGUACAAAGUAUGCUUUCAAUCAGG---CACUGAAUUGAAAAUCCAUUAACGUGUACGACG----- ---((((....))))...(((((((.(((((..(((--------((..---.)))))..))))).((((((((((.---..))).)))))))..........)))))))...----- ( -24.90, z-score = -1.56, R) >droPer1.super_2 3770397 95 - 9036312 ---CUGUAAACACAGUGCCGUACAUACGUACAGUGU--------ACGC---CGUACAAAGUAUGCUUUCAAUCAGG---CACUGAAUUGAAAAUCCAUUAACGUGUACGACG----- ---((((....))))...(((((((.(((((..(((--------((..---.)))))..))))).((((((((((.---..))).)))))))..........)))))))...----- ( -24.90, z-score = -1.56, R) >droWil1.scaffold_180701 3799405 102 + 3904529 ---------CCGCCUUUACCCUCUCCCGUUCUCUGU------CAACCAACUCAUACAAAGUAUGCUUUCAAUCAGGCAACACUGAAUUGAAAAUCCAUUAACGAGUACAAGGCAGAG ---------.....................((((((------(.....((((((((...))))..((((((((((......))).)))))))..........))))....))))))) ( -21.20, z-score = -2.74, R) >droMoj3.scaffold_6496 9734503 88 - 26866924 ---------------GUACUCACACACUUGCAU-----------ACACUGCAGUACAAAGUAUGCUUUCAAUCAGG---CGCUGAAUUGAAAAUCCAUUAACGAGUACAUAGUGCAG ---------------((((((........((((-----------((..((.....))..))))))((((((((((.---..))).)))))))..........))))))......... ( -22.10, z-score = -2.17, R) >consensus ________CACACAGCCACACACACACCUACAUCC_________ACACC__CACACAAAGUGUGCUUUCAAUCAGG___CACUGAAUUGAAAAUCCAUUAACGAGUACAAGGCGGAG ...................................................(((((...))))).((((((((((......))).)))))))......................... (-11.13 = -10.52 + -0.60)

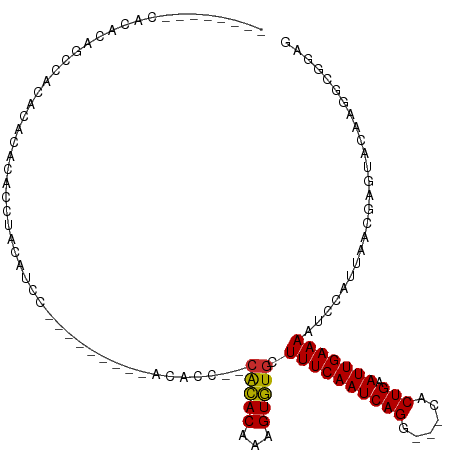

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:18 2011