| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,110,694 – 17,110,795 |
| Length | 101 |
| Max. P | 0.627490 |

| Location | 17,110,694 – 17,110,795 |
|---|---|
| Length | 101 |
| Sequences | 14 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 84.67 |
| Shannon entropy | 0.34978 |
| G+C content | 0.60598 |
| Mean single sequence MFE | -43.26 |
| Consensus MFE | -29.97 |
| Energy contribution | -31.81 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.627490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17110694 101 + 21146708 UCCUUGGUGGCCAGCAGCUGGUGCUGGCUG-UCGAUUUGCUGCUGCUGUCUCAAUGCCAUCGCCCUCAGUUCGGCUGACGUCAGCCUGCUGGGCACCUGCAA------------ .....((((.((((((...((((.((((..-..(((..((....)).))).....)))).))))........(((((....))))))))))).)))).....------------ ( -43.70, z-score = -1.50, R) >droPer1.super_2 3769583 102 + 9036312 UCCUUGGUGGCCAGCAGCUGGUGCUGGCUGGUCGAUUUGCUGCUGCUGUCUCAAGGCCAUCGCCCUCAGUUCGGCUGACGUCAGCCUGCUGGGCACCUGGAA------------ (((..((((.((((((...((((.(((((....(((..((....)).)))....))))).))))........(((((....))))))))))).)))).))).------------ ( -49.70, z-score = -2.27, R) >dp4.chr3 3580942 101 + 19779522 UCCUUGGUGGCCAGCAGCUGGUGCUGGCUG-UCGAUUUGCUGCUGCUGUCUCAAGGCCAUCGCCCUCAGUUCGGCUGACGUCAGCCUGCUGGGCACCUGGAA------------ (((..((((.((((((...((((.(((((.-..(((..((....)).)))....))))).))))........(((((....))))))))))).)))).))).------------ ( -49.30, z-score = -2.52, R) >droAna3.scaffold_13266 4901230 101 + 19884421 UCCUUGGUGGCCAGCAGUUGGUGCUGGCUG-UCGAUUUGCUGCUGCUGUCGCAAUGCCAUCGCCCUCAGUUCGGCUGACGUCAGCCUGCUGGGCACCUGGAA------------ (((..((((.(((((((..((((.((((((-.((((..((....)).))))))..)))).))))..).....(((((....))))))))))).)))).))).------------ ( -51.20, z-score = -3.56, R) >droEre2.scaffold_4845 11254268 101 + 22589142 UCCUUGGUGGCCAGCAGCUGGUGCUGGCUG-UCGAUUUGCUGCUGCUGUCUCAAUGCCAUCGCCCUCAGUUCGGCUGACGUCAGCCUGCUGGGCACCUGCAA------------ .....((((.((((((...((((.((((..-..(((..((....)).))).....)))).))))........(((((....))))))))))).)))).....------------ ( -43.70, z-score = -1.50, R) >droYak2.chr2R 9060025 101 - 21139217 UCCUUGGUGGCCAGCAGCUGGUGCUGGCUG-UCGAUUUGCUGCUGCUGUCUCAAUGCCAUCGCCCUCAGUUCGGCUGACGUCAGCCUGCUGGGCACCUGCAA------------ .....((((.((((((...((((.((((..-..(((..((....)).))).....)))).))))........(((((....))))))))))).)))).....------------ ( -43.70, z-score = -1.50, R) >droSec1.super_9 436935 101 + 3197100 UCCUUGGUGGCCAGCAGCUGGUGCUGGCUG-UCGAUUUGCUGCUGCUGUCUCAAAGCCAUCGCCCUCAGUUCGGCUGACGUCAGCCUGCUGGGCACCUGCAA------------ .....((((.((((((...((((.(((((.-..(((..((....)).)))....))))).))))........(((((....))))))))))).)))).....------------ ( -45.70, z-score = -2.01, R) >droSim1.chr2R 15761660 101 + 19596830 UCCUUGGUGGCCAGCAGCUGGUGCUGGCUG-UCGAUUUGCUGCUGCUGUCUCAAUGCCAUCGCCCUCAGUUCGGCUGACGUCAGCCUGCUGGGCACCUGCAA------------ .....((((.((((((...((((.((((..-..(((..((....)).))).....)))).))))........(((((....))))))))))).)))).....------------ ( -43.70, z-score = -1.50, R) >droMoj3.scaffold_6496 9732041 101 + 26866924 UCCUUUGUGGCCAGCAGCUGGUGCUGGCUG-UCGAUUUGCUGCUGCUGUCGCAAUGCCAUCGCCCUCAACUCGGCUGACGUCAGCCUGCUGGGCACCUGCAA------------ ......(((.((((((...((((.((((((-.((((..((....)).))))))..)))).))))........(((((....))))))))))).)))......------------ ( -43.00, z-score = -1.69, R) >droVir3.scaffold_12875 14284838 99 + 20611582 UCCUUUGUGGCCAGCAGCUGGUGCUGGCUG-UCGAUUUGCUGCUGCUGUCGCAGUGCCAUCGCCCUCAGUUCGGCUGACGUCAGCCUGCUGGGCACCUGC-------------- ......(((.(((((((.....((((((.(-(((((..((.(((((....))))))).)))(((........))).)))))))))))))))).)))....-------------- ( -43.10, z-score = -0.90, R) >droWil1.scaffold_180701 3798179 101 - 3904529 UCCUUUGUGGCCAGCAGCUGGUGCUGGCUG-UCGAUUUGCUGCUGCUGUCGCAAAGCCAUCGCCCUCAGUUCGGCUGACGUCAGCCUGCUGGGCACCUGCCA------------ ......(((.((((((...((((.(((((.-.((((..((....)).))))...))))).))))........(((((....))))))))))).)))......------------ ( -44.80, z-score = -1.57, R) >triCas2.ChLG4 11036513 107 + 13894384 UCUUUCGCGGCCAGCAGCUGGUGCUGCGAG-UCGAUCUGCGCUUGUUGUCUGCAGGCCAUCUCGCGCAGUUCGCUUAAUGUCAACUGCACACCGUCCGGUAACUGCAA------ ......((((..(.(.(.(((((..(((((-..(..((((((.....))..))))..)..)))))((((((.((.....)).)))))).))))).).).)..))))..------ ( -38.50, z-score = -0.68, R) >anoGam1.chrU 19169044 85 - 59568033 UCCUUUGCCGCAAGCAUCUGGUGCUGAGUA-UCAAUCUGAUGUUGUUGACGCAUUGCCAUUGAACGUAGUUCUGCAGCAGUCGGUC---------------------------- ......(((((.((((((((((((...)))-)))....))))))((((..(.(((((........))))).)..)))).).)))).---------------------------- ( -18.90, z-score = 1.32, R) >droGri2.scaffold_15245 14257652 113 + 18325388 UCCUUUGUGGCCAGCAGCUGGUGCUGGCUG-UCGAUUUGCUGCUGCUGUCGCAGUGCCAUCGCCCUCAGUUCGGCUGACGUCAGCCUGCUGGGCACCUGCAUAUAUGAGAGGGA ((((((..((((((((.....)))))))).-(((((.(((.(((((....)))))......((((..(((..(((((....))))).)))))))....))).)).))))))))) ( -46.60, z-score = -0.32, R) >consensus UCCUUGGUGGCCAGCAGCUGGUGCUGGCUG_UCGAUUUGCUGCUGCUGUCUCAAUGCCAUCGCCCUCAGUUCGGCUGACGUCAGCCUGCUGGGCACCUGCAA____________ ......(((.((((((...((((.((((.....(((..((....)).))).....)))).))))........(((((....))))))))))).))).................. (-29.97 = -31.81 + 1.84)

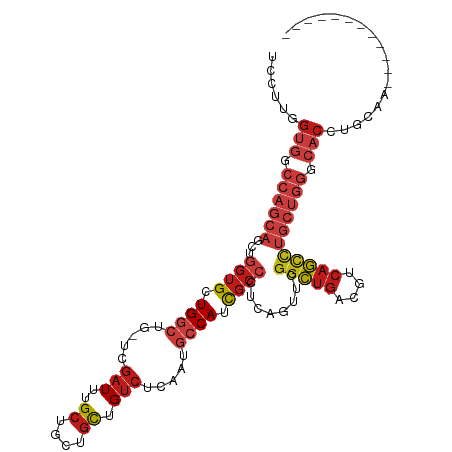
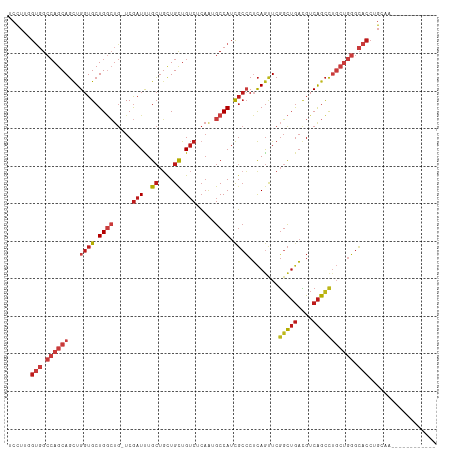
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:17 2011