| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,110,259 – 17,110,349 |
| Length | 90 |
| Max. P | 0.632943 |

| Location | 17,110,259 – 17,110,349 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 70.62 |
| Shannon entropy | 0.49594 |
| G+C content | 0.57691 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -19.77 |
| Energy contribution | -20.25 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.35 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.632943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
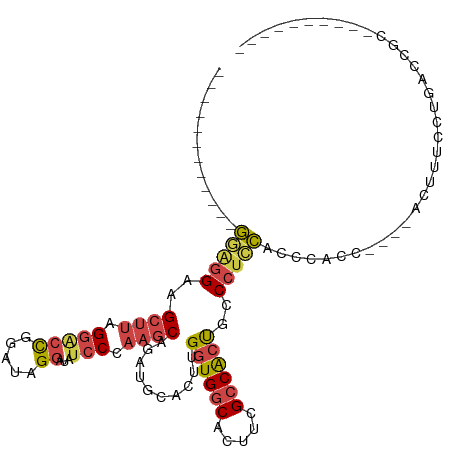
>dm3.chr2R 17110259 90 + 21146708 ------------GGAGGAAGCUUAGGGCCGGAUAGGAUAUCCCAAGCAGAUGCACUUGGUGGCACUUCGCCACUGCCCUCCACUCACC----ACUUUCCUGACCGC---------- ------------(((((..((((.(((((.....))....)))))))..........((((((.....))))))..))))).......----..............---------- ( -28.50, z-score = -0.64, R) >droSim1.chr2R 15761087 94 + 19596830 ------------GGAGGAAGCUUAGGACCGGAUAGGAUAUCCCAAGCAGAUGCACUUGGUGGCACUUCGCCGCUGCCCUCCACCCACCCAACACUUUCCUGACCGC---------- ------------(((((..((((.(((((.....))...))).))))..........((((((.....))))))..))))).........................---------- ( -27.70, z-score = -0.73, R) >droSec1.super_9 436397 94 + 3197100 ------------GGAGGAAGCUUAGGACCGGAUAGGAUAUCCCAAGCAGAUGCACUUGGUGGCACUUCGCCGCUGCCCUCCACCCACCCAACACUUUCCUGACCGC---------- ------------(((((..((((.(((((.....))...))).))))..........((((((.....))))))..))))).........................---------- ( -27.70, z-score = -0.73, R) >droYak2.chr2R 9059568 90 - 21139217 ------------GAGGGAAGCUUAGGAGCGGAUGGGAUAUCCCAAGCAGAUGCACUUGGUGGCACUUCGCCACCGGCCUUCAUCCACC----AUUUUCCUACCCAC---------- ------------..(((.....(((((((...((((....)))).)).((((...((((((((.....))))))))....))))....----....))))))))..---------- ( -32.50, z-score = -2.12, R) >droEre2.scaffold_4845 11253606 95 + 22589142 ------------GAGGGGAGCUUAGGACCGGAUGGGAUAUCCCAAGCAGAUGCACUUGGUGGCACUUCGCCACCGGCCUCCAACCGCC----AUUUUCCAACAUUUUUUGC----- ------------..((((.....(((.((...((((....)))).((....))....((((((.....)))))))))))....)).))----...................----- ( -28.90, z-score = -0.35, R) >droAna3.scaffold_13266 4900681 115 + 19884421 GAGGGGAGGUGUAGGAGUGGGAAGGCUCUGGAAAGGAUAUCCCAAGCCGAUGCACUCGUUGCCACUUUGCCACUAACC-CUGACCACCCUCCAGUCGCUUUGAUACUUGAAAGCCC (((((..(((.(((((((((.((((((..(((.......)))..))))...(((.....)))....)).)))))...)-)))))).))))).....(((((.(....).))))).. ( -38.70, z-score = -0.78, R) >consensus ____________GGAGGAAGCUUAGGACCGGAUAGGAUAUCCCAAGCAGAUGCACUUGGUGGCACUUCGCCACUGCCCUCCACCCACC____ACUUUCCUGACCGC__________ ............(((((..((((.(((((.....))...))).))))..........((((((.....))))))..)))))................................... (-19.77 = -20.25 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:16 2011