| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,106,413 – 17,106,531 |
| Length | 118 |
| Max. P | 0.513100 |

| Location | 17,106,413 – 17,106,531 |
|---|---|
| Length | 118 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.26 |
| Shannon entropy | 0.46424 |
| G+C content | 0.54790 |
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -20.55 |
| Energy contribution | -21.07 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.67 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.513100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
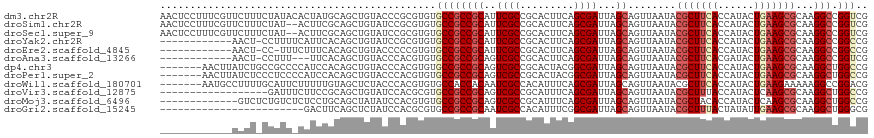
>dm3.chr2R 17106413 118 - 21146708 AACUCCUUUCGUUCUUUCUAUACACUAUGCAGCUGUACCCGCGUGUGCCGCCGCAUUCGCCGCACUUCAGCGAUUAGCAGUUAAUACGCUUCACCAUACUGAAGCGCAAGGCCGGUCG ........(((..(((..............((((((....(((.((((....)))).)))(((......)))....))))))....(((((((......))))))).)))..)))... ( -30.50, z-score = -0.57, R) >droSim1.chr2R 15757250 116 - 19596830 AACUCCUUUCGUUCUUUCUAU--ACUUCGCAGCUGUAUCCGCGUGUGCCGCCGCAUUCGCCGCACUUCAGCGAUUAGCAGUUAAUACGCUUCACCAUACUGAAGCGCAAGGCCGGUCG ........(((..(((.....--.......((((((....(((.((((....)))).)))(((......)))....))))))....(((((((......))))))).)))..)))... ( -30.50, z-score = -0.78, R) >droSec1.super_9 432642 116 - 3197100 AACUCCUUUCGUUCUUUCUAU--ACUUCGCAGCUGUAUCCGCGUGUGCCGCCGCAUUCGCCGCACUUCAGCGAUUAGCAGUUAAUACGCUUCACCAUACUGAAGCGCAAGGCCGGUCG ........(((..(((.....--.......((((((....(((.((((....)))).)))(((......)))....))))))....(((((((......))))))).)))..)))... ( -30.50, z-score = -0.78, R) >droYak2.chr2R 9055670 105 + 21139217 ------------AACU-CCUUUUCAUUCACAGCUGUAUCCGCGUGUGCCGCCGCAUUCGCCGCACUUCAGCGAUUAGCAGUUAAUACGCUUCACCAUACUGAAGCGCAAGGCCGGCCG ------------....-((((.........((((((....(((.((((....)))).)))(((......)))....))))))....(((((((......))))))).))))....... ( -28.60, z-score = -0.94, R) >droEre2.scaffold_4845 11249739 104 - 22589142 ------------AACU-CC-UUUCUUUCACAGCUGUACCCCCGUGUGCCGCCGCAUUCGCCGCACUUCAGCGAUUAGCAGUUAAUACGCUUCACCAUACUGAAGCGCAAGGCCGGCCG ------------....-..-........................(.((((((((..((((.........))))...))........(((((((......)))))))...)).))))). ( -25.40, z-score = -0.65, R) >droAna3.scaffold_13266 4896826 102 - 19884421 ------------AACU-CCUUU---UUCACAGCUGUACCCACGUGUGCCGCCGCAGUCGCCGCACUUCAGCGAUUAGCAGUUAAUACGCUUCACGAUACUGAAGCGCAAGGCCGGUCG ------------....-.....---..((((..((....))..))))(((((((((((((.........)))))).))........(((((((......)))))))...)).)))... ( -26.20, z-score = 0.15, R) >dp4.chr3 3577037 111 - 19779522 -------AACUUAUCUGCCGCCCCAUCCACAGCUGUACCCACGUGUGCCGCCGCAGUCGCCGCACUACGGCGAUUAGCAGUUAAUACGCUUCACCAUACUGAAGCGCAAGGCUGGCCG -------.........((((((.........((.((((......)))).)).((((((((((.....)))))))).))........(((((((......)))))))...))).))).. ( -38.00, z-score = -2.21, R) >droPer1.super_2 3765657 111 - 9036312 -------AACUUAUCUCCCUCCCCAUCCACAGCUGUACCCACGUGUGCCGCCGCAGUCGCCGCACUACGGCGAUUAGCAGUUAAUACGCUUCACCAUACUGAAGCGCAAGGCUGGCCG -------.....................................(.((((((((((((((((.....)))))))).))........(((((((......)))))))...))).)))). ( -34.60, z-score = -2.48, R) >droWil1.scaffold_180701 3792949 111 + 3904529 -------AAUGCCUUUUGCAUUCUUUUUGUAGCUCUACCCACGUGUGCCACCACAAUCGCCACAUUUCAGCGAUUAGCAGUUAAUACGCUUCACCAUACUGAAGAAAAAGGCCGGACG -------...(((((((............(((((((......(((......)))((((((.........)))))))).))))).....(((((......))))).)))))))...... ( -22.50, z-score = -0.85, R) >droVir3.scaffold_12875 14280113 100 - 20611582 ------------------GAUUUCUUCCGCAGCUGUAUCCACGCGUGCCGCCGCAGUCGCCGCAUUUCAGCGAUUAGCAGUUAAUACGCUUUACCAUACUCAAGCGCAAGGCUGGCCG ------------------.............((((....)).))(.((((((((((((((.........)))))).))........(((((..........)))))...))).)))). ( -23.70, z-score = 1.09, R) >droMoj3.scaffold_6496 9727199 105 - 26866924 -------------GUCUCUGUCUCUCCUGCAGCUAUAUCCACGUGUGCCGCCGCAGUCGCCGCAUUUCAGCGAUUAGCAGUUAAUACGCUACACCAUACUCAAGCGCAAGGCUGGCCG -------------(((...((((....(((.(((........((((((....((((((((.........)))))).)).((....)))).))))........)))))))))).))).. ( -23.39, z-score = 1.30, R) >droGri2.scaffold_15245 14252245 94 - 18325388 ------------------------GACUUCAGCUCUAUCCACGCGUGCCGCCGCAAUCGCCACAUUUCGGCGAUUAGCAGUUAAUACGCUUUACUAUAUUGAAGCGCAAGGCUGGGCG ------------------------.......((((...((..(((((..((.(((((((((.......))))))).)).))...)))((((((......))))))))..))..)))). ( -29.70, z-score = -1.38, R) >consensus ____________AACU_CCAUU_CUUUCGCAGCUGUACCCACGUGUGCCGCCGCAGUCGCCGCACUUCAGCGAUUAGCAGUUAAUACGCUUCACCAUACUGAAGCGCAAGGCCGGCCG ..............................................((((((((..((((.........))))...))........(((((((......)))))))...))).))).. (-20.55 = -21.07 + 0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:15 2011