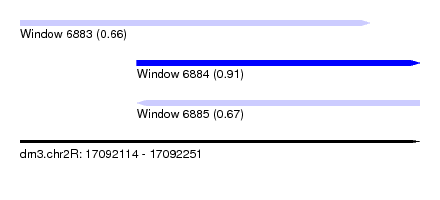
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,092,114 – 17,092,251 |
| Length | 137 |
| Max. P | 0.905291 |

| Location | 17,092,114 – 17,092,234 |
|---|---|
| Length | 120 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.95 |
| Shannon entropy | 0.43407 |
| G+C content | 0.59643 |
| Mean single sequence MFE | -46.69 |
| Consensus MFE | -28.22 |
| Energy contribution | -28.48 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.663818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
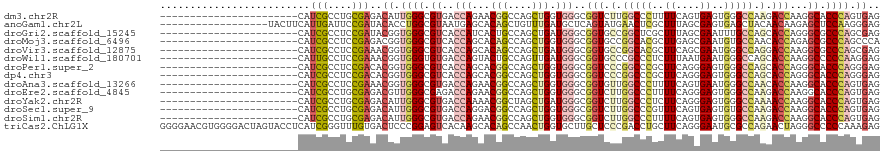
>dm3.chr2R 17092114 120 + 21146708 UAGAAUUCGCUUGGGUGACGCUGACGACAAAUGACACGUACUCACUGGGUGCCUUGGUCUUGGCCCACUCACUGAAAAGGGCCAAGACCGCCCACCAGCUGGCCGUUCUGGUCACGCCCA ...........((((((..((((..............(((((.....)))))...(((((((((((............)))))))))))......))))(((((.....))))))))))) ( -48.00, z-score = -2.61, R) >anoGam1.chr2L 48741587 120 + 48795086 CAGAAUACGCCUGGGUGACUUUAGCGACCAAUGAUUCCUACUCCCUUGGAGCUCUUGUUGUAGCUCACUCGCUAAAGCGAGUUCAUACUGAGCAUCAAACAGCUGUGCUCAUUACGCCAG ..........(((((((((((((((((.(((.(..........).)))((((((.....).)))))..)))))))))...........((((((.((......)))))))))))).)))) ( -32.50, z-score = -0.91, R) >droGri2.scaffold_15245 14229329 120 + 18325388 CAGAGUUCGCUUGGGUUACUUUGACCACAAAUGAUACGUACUCGCUGGGCGCCCUGGUGCUGGCAAAUUCGCUAAAGCGAGCCGGCACCGCCCAUCAGCUGGCAGUGAUGGUGACGCCCA ..(((((((.(((((((.....)))).))).))).....))))...((((((((.(((((((((....((((....)))))))))))))(((........)))......)).).))))). ( -50.00, z-score = -2.21, R) >droMoj3.scaffold_6496 9704808 120 + 26866924 UAGAGUUCGCCUGGGUUACCUUGACGACAAAUGAUAACUAUGGGCUGGGCGCUCUGGUGUUGGCACAUUCGCUCAAGCGUGCCGGCACCGCCCACCAGCUGGCUGUGCUGGUGACGCCCA ........((((((((((.(............).))))).))))).(((((....(((((((((((....((....)))))))))))))...(((((((.......))))))).))))). ( -51.10, z-score = -1.70, R) >droVir3.scaffold_12875 14249719 120 + 20611582 CAGAGUUCGCCUGGGUUACCCUGACCACAAAUGAUACCUACUCGCUGGGCGCCUUGGUCCUGGCCCAUUCGCUGAAGCGUGCCGGCACCGCCCAUCAGCUGGCUGUGCUGGUGACGCCCA ..(((((((..((((((.....)))).))..))).....))))...(((((....(((.(((((.....(((....))).))))).)))...(((((((.......))))))).))))). ( -43.70, z-score = -0.01, R) >droWil1.scaffold_180701 3773120 120 - 3904529 UAGAAUUCGCUUGGGUGACACUGACUACAAAUGACACGUACUCCUUGGGGGCCUUGGUGCUGGCCCAUUCAUUAAAGAGGGCGGGCACCGCCCAUCAACUGGCAGUACUGGUCACACCCA ...........((((((....((....))..((((..(((((((((((((((...((((((.((((..((......)))))).)))))))))).))))..)).)))))..)))))))))) ( -48.70, z-score = -2.85, R) >droPer1.super_2 3751226 120 + 9036312 CAGAGUUCGCUUGGGUGACGCUGACGACAAAUGACACGUACUCCCUGGGUGCCCUGGUGCUGGCCCACUCCCUGAAGCGGGCCGGGACCGCCCACCAGCUGGCCGUGCUGGUGACGCCCA ..(((....)))(((((..((................(((((.....)))))...(((.(((((((.((......)).))))))).))))).(((((((.......))))))).))))). ( -51.10, z-score = -0.52, R) >dp4.chr3 3562487 120 + 19779522 CAGAGUUCGCUUGGGUGACGCUGACGACAAAUGACACGUACUCCCUGGGUGCCCUGGUGCUGGCCCACUCCCUGAAGCGGGCCGGGACCGCCCACCAGCUGGCCGUGCUGGUGACGCCCA ..(((....)))(((((..((................(((((.....)))))...(((.(((((((.((......)).))))))).))))).(((((((.......))))))).))))). ( -51.10, z-score = -0.52, R) >droAna3.scaffold_13266 4871028 120 + 19884421 UAGAGUUCGCUUGGGUGACGCUGACGACAAAUGACACGUACUCACUGGGUGCCUUGGUGUUGGCCCAUUCACUGAAAAGGGCCAACACCGCCCACCAGCUGGCCGUUCUGGUCACGCCCA ..(((....)))(((((..((((..............(((((.....)))))...(((((((((((............)))))))))))......))))(((((.....)))))))))). ( -48.20, z-score = -1.98, R) >droEre2.scaffold_4845 11234663 120 + 22589142 UAGAGUUCGCUUGGGUGACGCUGACGACAAAUGACACGUACUCACUGGGUGCCUUGGUCUUGGCCCACUCCCUGAAAAGGGCCAAGACCGCCCACCAGCUGGCCGUUCUGGUCUCGCCCA ..(((....)))((((((.((((..............(((((.....)))))...(((((((((((............)))))))))))......)))).((((.....)))))))))). ( -48.30, z-score = -2.01, R) >droYak2.chr2R 9034945 120 - 21139217 CAGAGUUCGCUUGGGUGACGCUGACGACAAAUGACACGUACUCACUGGGUGCCUUGGUUUUGGCCCACUCCCUGAAGAGGGCCAAGACCGCCCAUCAGCUAGCCGUUUUGGUCACGCCCA ..(((....)))(((((..(((((.(...........(((((.....)))))...(((((((((((.((......)).)))))))))))...).)))))..(((.....)))..))))). ( -45.00, z-score = -1.50, R) >droSec1.super_9 418362 120 + 3197100 UAGAGUUCGCUUGGGUGACGCUGACGACAAAUGACACGUACUCACUGGGUGCCUUGGUCUUGGCACACUCACUGAAACGGGCCAAGACCGCCCACCAGCUGGCCGUCCUGGUCACGCCCA ..(((....)))(((((..((((..............(((((.....)))))...(((((((((.(............).)))))))))......))))(((((.....)))))))))). ( -41.40, z-score = -0.12, R) >droSim1.chr2R 15742441 120 + 19596830 CAGAGUUCGCUUGGGUGACGCUGACGACAAAUGACACGUACUCACUGGGUGCCUUGGUCUUGGCCCACUCACUGAAAAGGGCCAAGACCGCCCACCAGCUGGCCGUUCUGGUCACGCCCA ..(((....)))(((((..((((..............(((((.....)))))...(((((((((((............)))))))))))......))))(((((.....)))))))))). ( -48.40, z-score = -1.90, R) >triCas2.ChLG1X 7960892 120 - 8109244 UAGGUUUCGCGUGGGUGACUUUGGCCACGAACGACUCGUACUCUUUGGGGGCCCUAGUUCUGGCGCAUUCCCUGAAGCAGGUCGGGAGCAAGCACCAGUUGGCUGUGCUUGUGACUCCGG ..((..((((((.((.((((..((((((((.....)))).((.....))))))..)))))).))))....(((((......))))).((((((((.((....))))))))))))..)).. ( -46.20, z-score = -0.47, R) >consensus CAGAGUUCGCUUGGGUGACGCUGACGACAAAUGACACGUACUCACUGGGUGCCUUGGUGUUGGCCCACUCACUGAAGCGGGCCGAGACCGCCCACCAGCUGGCCGUGCUGGUCACGCCCA ............(((((((....(((...................(((((.....(((((((((((.((......)).))))))))))))))))((....)).)))....))))).)).. (-28.22 = -28.48 + 0.26)



| Location | 17,092,154 – 17,092,251 |
|---|---|
| Length | 97 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.57 |
| Shannon entropy | 0.47932 |
| G+C content | 0.64201 |
| Mean single sequence MFE | -41.57 |
| Consensus MFE | -27.67 |
| Energy contribution | -27.18 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.80 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
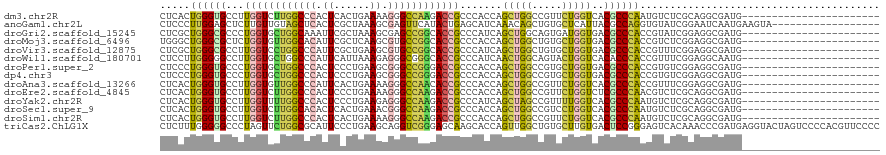
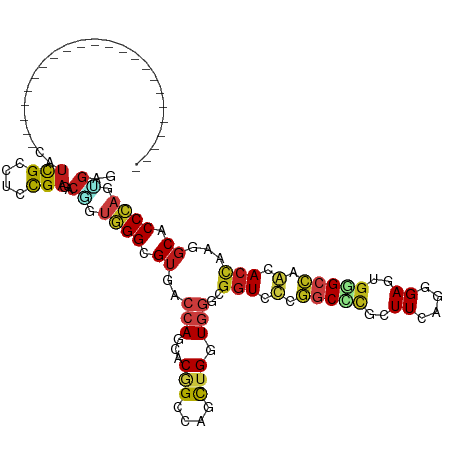
>dm3.chr2R 17092154 97 + 21146708 CUCACUGGGUGCCUUGGUCUUGGCCCACUCACUGAAAAGGGCCAAGACCGCCCACCAGCUGGCCGUUCUGGUCACGCCCAAUGUCUCGCAGGCGAUG----------------------- ..((.((((((....(((((((((((............)))))))))))((......))(((((.....))))))))))).))..(((....)))..----------------------- ( -39.70, z-score = -1.84, R) >anoGam1.chr2L 48741627 102 + 48795086 CUCCCUUGGAGCUCUUGUUGUAGCUCACUCGCUAAAGCGAGUUCAUACUGAGCAUCAAACAGCUGUGCUCAUUACGCCAGGUGUAUCGGAAUCAAUGAAGUA------------------ .(((.(((..((((.....(((....((((((....))))))...))).))))..)))(((.(((.((.......))))).)))...)))............------------------ ( -29.30, z-score = -1.19, R) >droGri2.scaffold_15245 14229369 97 + 18325388 CUCGCUGGGCGCCCUGGUGCUGGCAAAUUCGCUAAAGCGAGCCGGCACCGCCCAUCAGCUGGCAGUGAUGGUGACGCCCACCGUAUCGGAGGCGAUG----------------------- .(((((((((((((.(((((((((....((((....)))))))))))))(((........)))......)).).))))).(((...))).)))))..----------------------- ( -48.10, z-score = -2.62, R) >droMoj3.scaffold_6496 9704848 97 + 26866924 UGGGCUGGGCGCUCUGGUGUUGGCACAUUCGCUCAAGCGUGCCGGCACCGCCCACCAGCUGGCUGUGCUGGUGACGCCCACCGUCUCGGAGGCGAUG----------------------- ..((.((((((....(((((((((((....((....)))))))))))))...(((((((.......))))))).))))))))(((.....)))....----------------------- ( -49.60, z-score = -1.64, R) >droVir3.scaffold_12875 14249759 97 + 20611582 CUCGCUGGGCGCCUUGGUCCUGGCCCAUUCGCUGAAGCGUGCCGGCACCGCCCAUCAGCUGGCUGUGCUGGUGACGCCCACCGUUUCGGAGGCGAUG----------------------- .((((((((((....(((.(((((.....(((....))).))))).)))...(((((((.......))))))).))))).(((...))).)))))..----------------------- ( -43.40, z-score = -0.74, R) >droWil1.scaffold_180701 3773160 97 - 3904529 CUCCUUGGGGGCCUUGGUGCUGGCCCAUUCAUUAAAGAGGGCGGGCACCGCCCAUCAACUGGCAGUACUGGUCACACCCACCGUUUCGGAGGCAAUG----------------------- ((((.((((((....((((((.((((..((......)))))).))))))(((........))).............))).)))....))))......----------------------- ( -37.80, z-score = -0.74, R) >droPer1.super_2 3751266 97 + 9036312 CUCCCUGGGUGCCCUGGUGCUGGCCCACUCCCUGAAGCGGGCCGGGACCGCCCACCAGCUGGCCGUGCUGGUGACGCCCACCGUGUCGGAGGCGAUG----------------------- .((((((((((....(((.(((((((.((......)).))))))).)))...(((((((.......))))))).))))).(((...)))))).))..----------------------- ( -48.60, z-score = -1.16, R) >dp4.chr3 3562527 97 + 19779522 CUCCCUGGGUGCCCUGGUGCUGGCCCACUCCCUGAAGCGGGCCGGGACCGCCCACCAGCUGGCCGUGCUGGUGACGCCCACCGUGUCGGAGGCGAUG----------------------- .((((((((((....(((.(((((((.((......)).))))))).)))...(((((((.......))))))).))))).(((...)))))).))..----------------------- ( -48.60, z-score = -1.16, R) >droAna3.scaffold_13266 4871068 97 + 19884421 CUCACUGGGUGCCUUGGUGUUGGCCCAUUCACUGAAAAGGGCCAACACCGCCCACCAGCUGGCCGUUCUGGUCACGCCCACCGUUUCGGAGGCGAUG----------------------- .......((((...((((((((((((............))))))))))))..))))...(((((.....)))))((((..(((...))).))))...----------------------- ( -42.20, z-score = -2.20, R) >droEre2.scaffold_4845 11234703 97 + 22589142 CUCACUGGGUGCCUUGGUCUUGGCCCACUCCCUGAAAAGGGCCAAGACCGCCCACCAGCUGGCCGUUCUGGUCUCGCCCAACGUCUCGCAGGCGAUG----------------------- .....((((((....(((((((((((............)))))))))))((......)).((((.....)))).)))))).((((.....))))...----------------------- ( -39.90, z-score = -1.86, R) >droYak2.chr2R 9034985 97 - 21139217 CUCACUGGGUGCCUUGGUUUUGGCCCACUCCCUGAAGAGGGCCAAGACCGCCCAUCAGCUAGCCGUUUUGGUCACGCCCAAUGUCUCGCAGGCGAUG----------------------- ..((.((((((....(((((((((((.((......)).)))))))))))(.(((..(((.....))).))).).)))))).))..(((....)))..----------------------- ( -36.40, z-score = -1.27, R) >droSec1.super_9 418402 97 + 3197100 CUCACUGGGUGCCUUGGUCUUGGCACACUCACUGAAACGGGCCAAGACCGCCCACCAGCUGGCCGUCCUGGUCACGCCCAAUGUCUCGCAGGCGAUG----------------------- .(((.((((((((........))))).).)).)))....(((((.(((.(((........))).))).))))).((((...((.....))))))...----------------------- ( -33.50, z-score = 0.05, R) >droSim1.chr2R 15742481 97 + 19596830 CUCACUGGGUGCCUUGGUCUUGGCCCACUCACUGAAAAGGGCCAAGACCGCCCACCAGCUGGCCGUUCUGGUCACGCCCAAUGUCUCGCAGGCGAUG----------------------- ..((.((((((....(((((((((((............)))))))))))((......))(((((.....))))))))))).))..(((....)))..----------------------- ( -39.70, z-score = -1.84, R) >triCas2.ChLG1X 7960932 120 - 8109244 CUCUUUGGGGGCCCUAGUUCUGGCGCAUUCCCUGAAGCAGGUCGGGAGCAAGCACCAGUUGGCUGUGCUUGUGACUCCGGGAGUCACAAACCCGAUGAGGUACUAGUCCCCACGUUCCCC .....(((((((..(((((((...((.(((...)))))..((((((.((((((((.((....))))))))))(((((...))))).....)))))).))).)))))))))))........ ( -45.20, z-score = -0.52, R) >consensus CUCACUGGGUGCCUUGGUGUUGGCCCACUCACUGAAGCGGGCCGAGACCGCCCACCAGCUGGCCGUGCUGGUCACGCCCACCGUCUCGGAGGCGAUG_______________________ .....((((((...((((((((((((.((......)).)))))))))))).......(((((.....)))))..))))))........................................ (-27.67 = -27.18 + -0.49)
| Location | 17,092,154 – 17,092,251 |
|---|---|
| Length | 97 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.57 |
| Shannon entropy | 0.47932 |
| G+C content | 0.64201 |
| Mean single sequence MFE | -43.79 |
| Consensus MFE | -20.48 |
| Energy contribution | -20.81 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.665732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
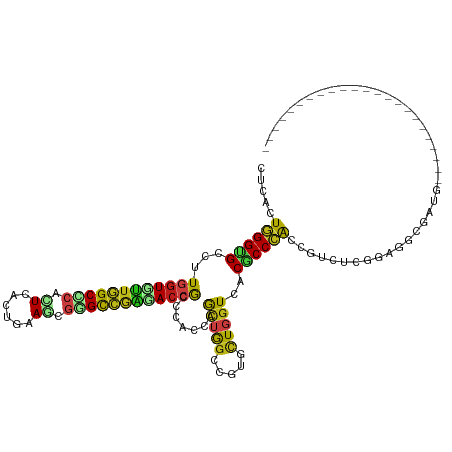
>dm3.chr2R 17092154 97 - 21146708 -----------------------CAUCGCCUGCGAGACAUUGGGCGUGACCAGAACGGCCAGCUGGUGGGCGGUCUUGGCCCUUUUCAGUGAGUGGGCCAAGACCAAGGCACCCAGUGAG -----------------------..(((....)))..((((((((((.......)))(((.(((....)))((((((((((((((.....))).)))))))))))..))).))))))).. ( -46.00, z-score = -2.68, R) >anoGam1.chr2L 48741627 102 - 48795086 ------------------UACUUCAUUGAUUCCGAUACACCUGGCGUAAUGAGCACAGCUGUUUGAUGCUCAGUAUGAACUCGCUUUAGCGAGUGAGCUACAACAAGAGCUCCAAGGGAG ------------------..((((.(((................((((.(((((((((....))).)))))).)))).((((((....))))))(((((........)))))))).)))) ( -30.10, z-score = -1.57, R) >droGri2.scaffold_15245 14229369 97 - 18325388 -----------------------CAUCGCCUCCGAUACGGUGGGCGUCACCAUCACUGCCAGCUGAUGGGCGGUGCCGGCUCGCUUUAGCGAAUUUGCCAGCACCAGGGCGCCCAGCGAG -----------------------.((((....)))).((.((((((((.((((((........))))))..(((((.(((((((....))))....))).)))))..)))))))).)).. ( -49.00, z-score = -3.35, R) >droMoj3.scaffold_6496 9704848 97 - 26866924 -----------------------CAUCGCCUCCGAGACGGUGGGCGUCACCAGCACAGCCAGCUGGUGGGCGGUGCCGGCACGCUUGAGCGAAUGUGCCAACACCAGAGCGCCCAGCCCA -----------------------..(((....)))...((((((((((((((((.......)))))))...((((..((((((.((....)).))))))..))))...))))))).)).. ( -43.50, z-score = -0.92, R) >droVir3.scaffold_12875 14249759 97 - 20611582 -----------------------CAUCGCCUCCGAAACGGUGGGCGUCACCAGCACAGCCAGCUGAUGGGCGGUGCCGGCACGCUUCAGCGAAUGGGCCAGGACCAAGGCGCCCAGCGAG -----------------------..((((..(((...)))((((((((.(((...(((....))).)))..(((.(.(((.(((....))).....))).).)))..)))))))))))). ( -41.90, z-score = -0.71, R) >droWil1.scaffold_180701 3773160 97 + 3904529 -----------------------CAUUGCCUCCGAAACGGUGGGUGUGACCAGUACUGCCAGUUGAUGGGCGGUGCCCGCCCUCUUUAAUGAAUGGGCCAGCACCAAGGCCCCCAAGGAG -----------------------......((((.....((.(((((.(....((((((((........)))))))))))))).)).........(((((........)))))....)))) ( -35.80, z-score = -0.55, R) >droPer1.super_2 3751266 97 - 9036312 -----------------------CAUCGCCUCCGACACGGUGGGCGUCACCAGCACGGCCAGCUGGUGGGCGGUCCCGGCCCGCUUCAGGGAGUGGGCCAGCACCAGGGCACCCAGGGAG -----------------------...((((((((...))).)))))((((((((.......))))))))....(((((((((((((....))))))))).......((....)).)))). ( -50.40, z-score = -1.32, R) >dp4.chr3 3562527 97 - 19779522 -----------------------CAUCGCCUCCGACACGGUGGGCGUCACCAGCACGGCCAGCUGGUGGGCGGUCCCGGCCCGCUUCAGGGAGUGGGCCAGCACCAGGGCACCCAGGGAG -----------------------...((((((((...))).)))))((((((((.......))))))))....(((((((((((((....))))))))).......((....)).)))). ( -50.40, z-score = -1.32, R) >droAna3.scaffold_13266 4871068 97 - 19884421 -----------------------CAUCGCCUCCGAAACGGUGGGCGUGACCAGAACGGCCAGCUGGUGGGCGGUGUUGGCCCUUUUCAGUGAAUGGGCCAACACCAAGGCACCCAGUGAG -----------------------....(((((((...((((((.(((.......))).))).))).)))..((((((((((((((.....))).))))))))))).)))).......... ( -44.30, z-score = -2.04, R) >droEre2.scaffold_4845 11234703 97 - 22589142 -----------------------CAUCGCCUGCGAGACGUUGGGCGAGACCAGAACGGCCAGCUGGUGGGCGGUCUUGGCCCUUUUCAGGGAGUGGGCCAAGACCAAGGCACCCAGUGAG -----------------------..(((((..(.(...(((((.((.........)).)))))).)..)))(((((((((((.(((....))).)))))))))))............)). ( -43.40, z-score = -1.59, R) >droYak2.chr2R 9034985 97 + 21139217 -----------------------CAUCGCCUGCGAGACAUUGGGCGUGACCAAAACGGCUAGCUGAUGGGCGGUCUUGGCCCUCUUCAGGGAGUGGGCCAAAACCAAGGCACCCAGUGAG -----------------------..(((....)))..((((((((((.......)))(((.(((....)))(((.(((((((.(((....))).))))))).)))..))).))))))).. ( -41.60, z-score = -2.00, R) >droSec1.super_9 418402 97 - 3197100 -----------------------CAUCGCCUGCGAGACAUUGGGCGUGACCAGGACGGCCAGCUGGUGGGCGGUCUUGGCCCGUUUCAGUGAGUGUGCCAAGACCAAGGCACCCAGUGAG -----------------------..(((....)))..((((((((((.......)))(((.(((....)))(((((((((.(((((....))))).)))))))))..))).))))))).. ( -39.20, z-score = -0.67, R) >droSim1.chr2R 15742481 97 - 19596830 -----------------------CAUCGCCUGCGAGACAUUGGGCGUGACCAGAACGGCCAGCUGGUGGGCGGUCUUGGCCCUUUUCAGUGAGUGGGCCAAGACCAAGGCACCCAGUGAG -----------------------..(((....)))..((((((((((.......)))(((.(((....)))((((((((((((((.....))).)))))))))))..))).))))))).. ( -46.00, z-score = -2.68, R) >triCas2.ChLG1X 7960932 120 + 8109244 GGGGAACGUGGGGACUAGUACCUCAUCGGGUUUGUGACUCCCGGAGUCACAAGCACAGCCAACUGGUGCUUGCUCCCGACCUGCUUCAGGGAAUGCGCCAGAACUAGGGCCCCCAAAGAG ((((..(.(((((.((.((.((.....))(((((((((((...)))))))))))))))))..(((((((....((((((......)).))))..)))))))..))).)..))))...... ( -51.40, z-score = -2.17, R) >consensus _______________________CAUCGCCUCCGAGACGGUGGGCGUGACCAGCACGGCCAGCUGGUGGGCGGUCCCGGCCCGCUUCAGGGAGUGGGCCAACACCAAGGCACCCAGUGAG .........................(((....)))..((.((((.((..(((...(((....))).)))..(((.(.(((((..((....))..))))).).)))...)).)))).)).. (-20.48 = -20.81 + 0.34)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:14 2011