| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,070,358 – 17,070,433 |
| Length | 75 |
| Max. P | 0.992199 |

| Location | 17,070,358 – 17,070,433 |
|---|---|
| Length | 75 |
| Sequences | 14 |
| Columns | 83 |
| Reading direction | forward |
| Mean pairwise identity | 79.28 |
| Shannon entropy | 0.49031 |
| G+C content | 0.61997 |
| Mean single sequence MFE | -33.54 |
| Consensus MFE | -27.32 |
| Energy contribution | -27.91 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.987718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
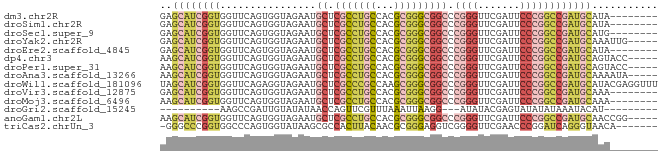
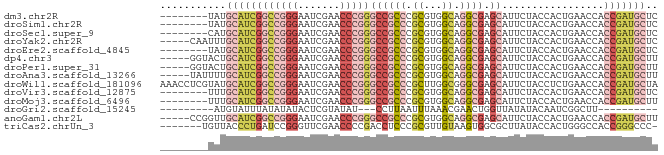
>dm3.chr2R 17070358 75 + 21146708 GAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUA-------- ..(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))...-------- ( -35.90, z-score = -1.86, R) >droSim1.chr2R 15719251 75 + 19596830 GAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUA-------- ..(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))...-------- ( -35.90, z-score = -1.86, R) >droSec1.super_9 396781 75 + 3197100 GAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUG-------- ..(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))...-------- ( -35.90, z-score = -1.87, R) >droYak2.chr2R 9013178 78 - 21139217 GAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAAUUG----- ..(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))......----- ( -35.90, z-score = -1.96, R) >droEre2.scaffold_4845 11212999 75 + 22589142 GAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUA-------- ..(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))...-------- ( -35.90, z-score = -1.86, R) >dp4.chr3 17721730 78 + 19779522 AAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAGUACC----- ..(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))......----- ( -35.90, z-score = -1.76, R) >droPer1.super_31 862815 78 - 935084 AAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAGUACC----- ..(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))......----- ( -35.90, z-score = -1.76, R) >droAna3.scaffold_13266 12990009 78 - 19884421 AAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAAAUA----- ..(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))......----- ( -35.90, z-score = -2.09, R) >droWil1.scaffold_181096 7895069 83 - 12416693 UAGCAUCGGUGGUUCAGAGGUAGAAUGCUCGCCCGCCAAGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUACGAGGUUU ..(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))........... ( -38.10, z-score = -2.19, R) >droVir3.scaffold_12875 10888530 75 + 20611582 GAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAA-------- ..(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))...-------- ( -35.90, z-score = -1.91, R) >droMoj3.scaffold_6496 10426576 75 - 26866924 AAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAA-------- ..(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))...-------- ( -35.90, z-score = -2.03, R) >droGri2.scaffold_15245 14334376 61 - 18325388 ----------AAGCCGAUUGUAUAUAACCAGUUCGUUUAAAUUAAGG---AUAUACGAGUAUAUAUAAAUACAU--------- ----------.......((((((((....((((......))))....---))))))))((((......))))..--------- ( -7.00, z-score = -0.43, R) >anoGam1.chr2L 33891052 78 - 48795086 AAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAACCGG----- ..(((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))......----- ( -35.90, z-score = -1.35, R) >triCas2.chrUn_3 246029 75 - 565036 -GGGCCCGGUGGCCCAGUGGUAUAAGCGCCACUUACAACGCGGGAGGUCGGGGUUCGAACCCGGAUCAGGGUAACA------- -(((((....)))))((((((......))))))....((.(....((((.((((....)))).)))).).))....------- ( -29.60, z-score = -0.52, R) >consensus GAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAA________ ..(((((....((((.(((((((.........)))))))..))))((((.(((......))).)))))))))........... (-27.32 = -27.91 + 0.59)
| Location | 17,070,358 – 17,070,433 |
|---|---|
| Length | 75 |
| Sequences | 14 |
| Columns | 83 |
| Reading direction | reverse |
| Mean pairwise identity | 79.28 |
| Shannon entropy | 0.49031 |
| G+C content | 0.61997 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -24.16 |
| Energy contribution | -25.33 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.992199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
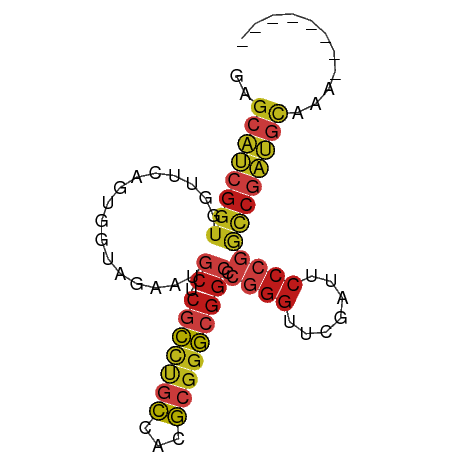
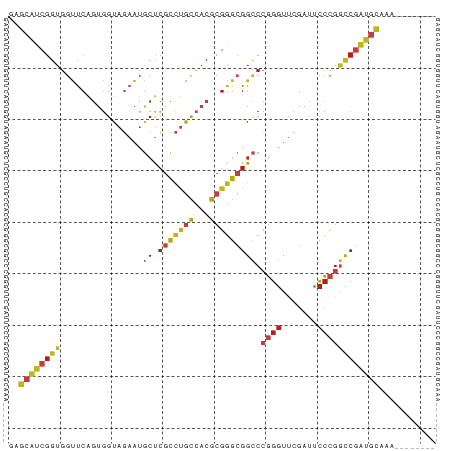
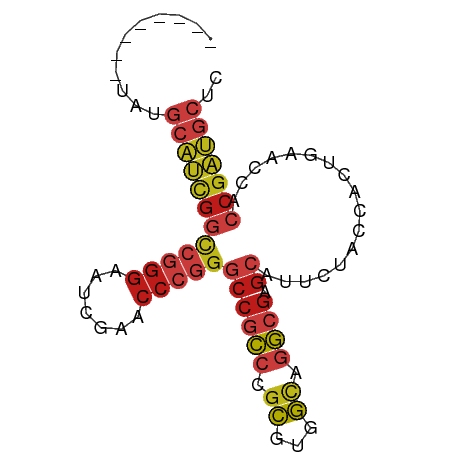
>dm3.chr2R 17070358 75 - 21146708 --------UAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUC --------...((((((((((((.......)))))((((((.((...)).)))).)).................))))))).. ( -32.20, z-score = -2.31, R) >droSim1.chr2R 15719251 75 - 19596830 --------UAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUC --------...((((((((((((.......)))))((((((.((...)).)))).)).................))))))).. ( -32.20, z-score = -2.31, R) >droSec1.super_9 396781 75 - 3197100 --------CAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUC --------...((((((((((((.......)))))((((((.((...)).)))).)).................))))))).. ( -32.20, z-score = -2.22, R) >droYak2.chr2R 9013178 78 + 21139217 -----CAAUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUC -----......((((((((((((.......)))))((((((.((...)).)))).)).................))))))).. ( -32.20, z-score = -2.28, R) >droEre2.scaffold_4845 11212999 75 - 22589142 --------UAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUC --------...((((((((((((.......)))))((((((.((...)).)))).)).................))))))).. ( -32.20, z-score = -2.31, R) >dp4.chr3 17721730 78 - 19779522 -----GGUACUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUU -----......((((((((((((.......)))))((((((.((...)).)))).)).................))))))).. ( -32.20, z-score = -1.59, R) >droPer1.super_31 862815 78 + 935084 -----GGUACUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUU -----......((((((((((((.......)))))((((((.((...)).)))).)).................))))))).. ( -32.20, z-score = -1.59, R) >droAna3.scaffold_13266 12990009 78 + 19884421 -----UAUUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUU -----......((((((((((((.......)))))((((((.((...)).)))).)).................))))))).. ( -32.20, z-score = -2.33, R) >droWil1.scaffold_181096 7895069 83 + 12416693 AAACCUCGUAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCUUGGCGGGCGAGCAUUCUACCUCUGAACCACCGAUGCUA ...........((((((((((((.......)))))(((((((((...))))))).)).................))))))).. ( -38.30, z-score = -3.48, R) >droVir3.scaffold_12875 10888530 75 - 20611582 --------UUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUC --------...((((((((((((.......)))))((((((.((...)).)))).)).................))))))).. ( -32.20, z-score = -2.28, R) >droMoj3.scaffold_6496 10426576 75 + 26866924 --------UUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUU --------...((((((((((((.......)))))((((((.((...)).)))).)).................))))))).. ( -32.20, z-score = -2.28, R) >droGri2.scaffold_15245 14334376 61 + 18325388 ---------AUGUAUUUAUAUAUACUCGUAUAU---CCUUAAUUUAAACGAACUGGUUAUAUACAAUCGGCUU---------- ---------..((((......))))((((.((.---........)).)))).((((((......))))))...---------- ( -6.00, z-score = -0.02, R) >anoGam1.chr2L 33891052 78 + 48795086 -----CCGGUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUU -----......((((((((((((.......)))))((((((.((...)).)))).)).................))))))).. ( -32.20, z-score = -1.17, R) >triCas2.chrUn_3 246029 75 + 565036 -------UGUUACCCUGAUCCGGGUUCGAACCCCGACCUCCCGCGUUGUAAGUGGCGCUUAUACCACUGGGCCACCGGGCCC- -------.........(((.((((.(((.....)))...)))).)))...(((((........)))))(((((....)))))- ( -23.80, z-score = 0.11, R) >consensus ________UAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUC ...........((((((((((((.......)))))((((((.((...)).)))).)).................))))))).. (-24.16 = -25.33 + 1.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:07 2011