
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,070,182 – 17,070,258 |
| Length | 76 |
| Max. P | 0.826085 |

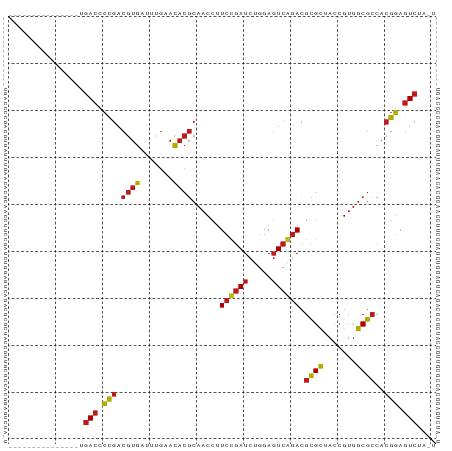
| Location | 17,070,182 – 17,070,258 |
|---|---|
| Length | 76 |
| Sequences | 14 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 84.69 |
| Shannon entropy | 0.32115 |
| G+C content | 0.57417 |
| Mean single sequence MFE | -22.54 |
| Consensus MFE | -19.83 |
| Energy contribution | -19.66 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.826085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
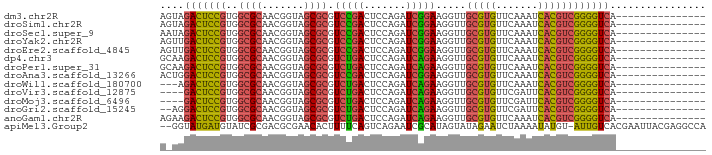
>dm3.chr2R 17070182 76 + 21146708 ---------------UGACCCCGACGUGAUUUGAACACGCAACCUUCCGAUCUGGAGUCGGACGCGCUACCGUUGCGCCACGGAGUCUACU ---------------.(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))).... ( -23.70, z-score = -1.42, R) >droSim1.chr2R 15719075 76 + 19596830 ---------------UGACCCCGACGUGAUUUGAACACGCAACCUUCCGAUCUGGAGUCGGACGCGCUACCGUUGCGCCACGGAGUCUACU ---------------.(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))).... ( -23.70, z-score = -1.42, R) >droSec1.super_9 396605 76 + 3197100 ---------------UGACCCCGACGUGAUUUGAACACGCAACCUUCCGAUCUGGAGUCGGACGCGCUACCGUUGCGCCACGGAGUCUAUU ---------------.(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))).... ( -23.70, z-score = -1.44, R) >droYak2.chr2R 9013002 76 - 21139217 ---------------UGACCCCGACGUGAUUUGAACACGCAACCUUCCGAUCUGGAGUCGGACGCGCUACCGUUGCGCCACGGAGUCAACU ---------------((((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))))... ( -24.90, z-score = -1.70, R) >droEre2.scaffold_4845 11212824 76 + 22589142 ---------------UGACCCCGACGUGAUUUGAACACGCAACCUUCCGAUCUGGAGUCGGACGCGCUACCGUUGCGCCACGGAGUCAACU ---------------((((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))))... ( -24.90, z-score = -1.70, R) >dp4.chr3 17721550 76 + 19779522 ---------------UGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCUUGC ---------------.(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))).... ( -22.20, z-score = -1.07, R) >droPer1.super_31 862635 76 - 935084 ---------------UGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCUUGC ---------------.(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))).... ( -22.20, z-score = -1.07, R) >droAna3.scaffold_13266 12989830 76 - 19884421 ---------------UGACCCCGACGUGAUUUGAACACGCAACCUUCCGAUCUGGAGUCGGACGCGCUACCGUUGCGCCACGGAGUCCAGU ---------------.(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))).... ( -23.30, z-score = -0.65, R) >droWil1.scaffold_180700 4811158 73 + 6630534 ---------------UGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCU--- ---------------.(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))).--- ( -22.20, z-score = -1.46, R) >droVir3.scaffold_12875 9724045 72 - 20611582 ---------------UGACCCCGACGUGAAUCGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUC---- ---------------.(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).)))---- ( -21.00, z-score = -0.77, R) >droMoj3.scaffold_6496 16050184 72 + 26866924 ---------------UGACCCCGACGUGAAUCGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUC---- ---------------.(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).)))---- ( -21.00, z-score = -0.77, R) >droGri2.scaffold_15245 14334219 74 - 18325388 ---------------UGACCCCGACGUGAAUCGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCCU-- ---------------.(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).)))..-- ( -21.80, z-score = -1.13, R) >anoGam1.chr2R 57345791 76 + 62725911 ---------------UGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCUUCU ---------------.(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))).... ( -22.20, z-score = -1.25, R) >apiMel3.Group2 7681465 88 - 14465785 UGGCCUCGUAAUUCGUGACAAU-ACAUAUUUUAGAUUCUAUACUAUGCGAUUCUGACUGAAAAGUGUUCGCGUCGCGAUACAUCAUACC-- .......(((..(((((((...-........(((........))).((((.....(((....)))..))))))))))))))........-- ( -18.70, z-score = -0.86, R) >consensus _______________UGACCCCGACGUGAUUUGAACACGCAACCUUCCGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCUA_U ................(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))).... (-19.83 = -19.66 + -0.17)


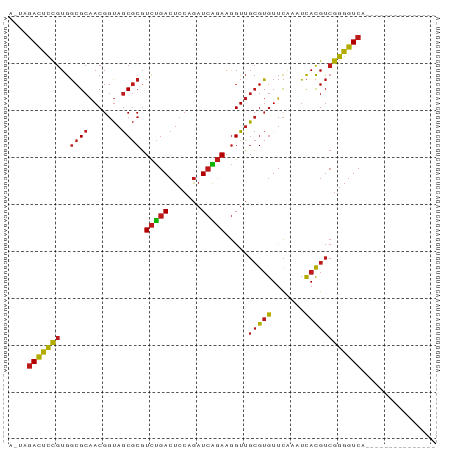
| Location | 17,070,182 – 17,070,258 |
|---|---|
| Length | 76 |
| Sequences | 14 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 84.69 |
| Shannon entropy | 0.32115 |
| G+C content | 0.57417 |
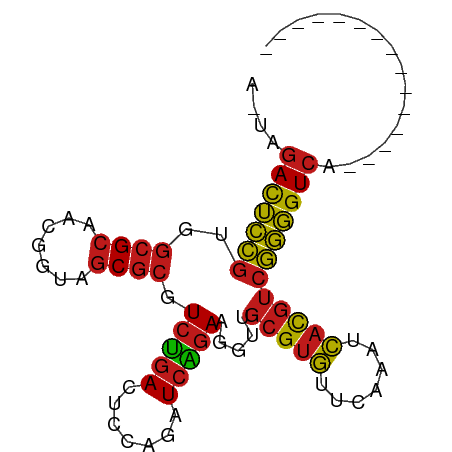
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -24.03 |
| Energy contribution | -23.59 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.781309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17070182 76 - 21146708 AGUAGACUCCGUGGCGCAACGGUAGCGCGUCCGACUCCAGAUCGGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCA--------------- ....(((((((..((((.......)))).((((((....).))))).....(((((.......)))))))))))).--------------- ( -28.10, z-score = -1.30, R) >droSim1.chr2R 15719075 76 - 19596830 AGUAGACUCCGUGGCGCAACGGUAGCGCGUCCGACUCCAGAUCGGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCA--------------- ....(((((((..((((.......)))).((((((....).))))).....(((((.......)))))))))))).--------------- ( -28.10, z-score = -1.30, R) >droSec1.super_9 396605 76 - 3197100 AAUAGACUCCGUGGCGCAACGGUAGCGCGUCCGACUCCAGAUCGGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCA--------------- ....(((((((..((((.......)))).((((((....).))))).....(((((.......)))))))))))).--------------- ( -28.10, z-score = -1.52, R) >droYak2.chr2R 9013002 76 + 21139217 AGUUGACUCCGUGGCGCAACGGUAGCGCGUCCGACUCCAGAUCGGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCA--------------- ...((((((((..((((.......)))).((((((....).))))).....(((((.......)))))))))))))--------------- ( -28.00, z-score = -1.01, R) >droEre2.scaffold_4845 11212824 76 - 22589142 AGUUGACUCCGUGGCGCAACGGUAGCGCGUCCGACUCCAGAUCGGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCA--------------- ...((((((((..((((.......)))).((((((....).))))).....(((((.......)))))))))))))--------------- ( -28.00, z-score = -1.01, R) >dp4.chr3 17721550 76 - 19779522 GCAAGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCA--------------- ....(((((((..((((.......)))).((((((....).))))).....(((((.......)))))))))))).--------------- ( -26.60, z-score = -1.20, R) >droPer1.super_31 862635 76 + 935084 GCAAGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCA--------------- ....(((((((..((((.......)))).((((((....).))))).....(((((.......)))))))))))).--------------- ( -26.60, z-score = -1.20, R) >droAna3.scaffold_13266 12989830 76 + 19884421 ACUGGACUCCGUGGCGCAACGGUAGCGCGUCCGACUCCAGAUCGGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCA--------------- ...((((......((((.......))))))))((((((.((((....))))(((((.......))))).)))))).--------------- ( -29.80, z-score = -1.24, R) >droWil1.scaffold_180700 4811158 73 - 6630534 ---AGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCA--------------- ---.(((((((..((((.......)))).((((((....).))))).....(((((.......)))))))))))).--------------- ( -26.60, z-score = -1.47, R) >droVir3.scaffold_12875 9724045 72 + 20611582 ----GACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCGAUUCACGUCGGGGUCA--------------- ----(((((((..((((.......)))).((((((....).))))).....(((((.......)))))))))))).--------------- ( -26.40, z-score = -0.82, R) >droMoj3.scaffold_6496 16050184 72 - 26866924 ----GACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCGAUUCACGUCGGGGUCA--------------- ----(((((((..((((.......)))).((((((....).))))).....(((((.......)))))))))))).--------------- ( -26.40, z-score = -0.82, R) >droGri2.scaffold_15245 14334219 74 + 18325388 --AGGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCGAUUCACGUCGGGGUCA--------------- --..(((((((..((((.......)))).((((((....).))))).....(((((.......)))))))))))).--------------- ( -26.40, z-score = -0.60, R) >anoGam1.chr2R 57345791 76 - 62725911 AGAAGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCA--------------- ....(((((((..((((.......)))).((((((....).))))).....(((((.......)))))))))))).--------------- ( -26.60, z-score = -1.24, R) >apiMel3.Group2 7681465 88 + 14465785 --GGUAUGAUGUAUCGCGACGCGAACACUUUUCAGUCAGAAUCGCAUAGUAUAGAAUCUAAAAUAUGU-AUUGUCACGAAUUACGAGGCCA --(((....(((((((.(((((((......(((.....)))))))..((((((............)))-)))))).)))..))))..))). ( -16.70, z-score = 0.38, R) >consensus A_UAGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCA_______________ ....(((((((..((((.......)))).(((((.......))))).....(((((.......))))))))))))................ (-24.03 = -23.59 + -0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:05 2011