| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,068,465 – 17,068,557 |
| Length | 92 |
| Max. P | 0.998819 |

| Location | 17,068,465 – 17,068,557 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 68.72 |
| Shannon entropy | 0.60335 |
| G+C content | 0.46255 |
| Mean single sequence MFE | -24.89 |
| Consensus MFE | -13.19 |
| Energy contribution | -15.33 |
| Covariance contribution | 2.14 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.50 |
| SVM RNA-class probability | 0.998819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
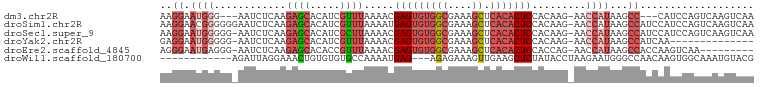
>dm3.chr2R 17068465 92 + 21146708 AAGGAAUGGG---AAUCUCAAGAGCACAUCGUUUAAAACGAGUGUGGCGAAAGCUCACACUCCACAAG-AACCAUAAGCC---CAUCCAGUCAAGUCAA ..(((.((((---........((.....))((((.....(((((((((....)).))))))).....)-)))......))---)))))........... ( -28.00, z-score = -3.98, R) >droSim1.chr2R 15717359 98 + 19596830 AAGGAACGGGGGGAAUCUCAAGAGCACAUCGUUUAAAAUGAGUGUGGCGAAAGCUCACACUCCACAAG-AACCAUAAGCCAUCCAUCCAGUCAAGUCAA ...((..((..(((.((....))((..((.((((.....(((((((((....)).))))))).....)-))).))..))..)))..))..))....... ( -26.90, z-score = -2.25, R) >droSec1.super_9 394906 97 + 3197100 AAGGAAUGGGGG-AAUCUCAAGAGCACAUCGCUUAAAACGAGUGUGGCGAAAGCUCACACUCCACAAG-AACCAUAAGCCAUCCAUCCAGUCAAGUCAA ..(((.((((((-........((((.....)))).....(((((((((....)).)))))))......-.........)).)))))))........... ( -30.00, z-score = -3.52, R) >droYak2.chr2R 9011332 83 - 21139217 GAGGAAUGGGGG-AAUCUCAAGAGCACAUCGUUUAAAACGAGUGUGGCGAAAGCUCACACUCCACAAG-AACCAUAAGCCAUCAA-------------- ((((.((((...-........((((.....)))).....(((((((((....)).)))))))......-..))))...)).))..-------------- ( -24.60, z-score = -2.91, R) >droEre2.scaffold_4845 11211123 88 + 22589142 AGGGAAUGAGGG-AAUCUCAAGAGCACACCGUUUAAAACGAGUGUGGCGAAAGCUCACACUCCACCAG-AACCAUAAGCCACCAAGUCAA--------- .((...((((..-...)))).(.((.....((((.....(((((((((....)).))))))).....)-))).....))).)).......--------- ( -24.30, z-score = -2.21, R) >droWil1.scaffold_180700 4809228 84 + 6630534 ------------AGAUUAGGAAACUGUGUGUGCCAAAAUGAG---AGAGAAAGUUGAAGCUCUAUACCUAAGAAUGGGCCAACAAGUGGCAAAUGUACG ------------.....((....))(((..(((((...(...---.).....((((..((((.............))))))))...))))..)..))). ( -15.52, z-score = -0.43, R) >consensus AAGGAAUGGGGG_AAUCUCAAGAGCACAUCGUUUAAAACGAGUGUGGCGAAAGCUCACACUCCACAAG_AACCAUAAGCCAUCAAUCCAGUCAAGUCAA ..((.((((............((((.....)))).....(((((((((....)).))))))).........))))...))................... (-13.19 = -15.33 + 2.14)
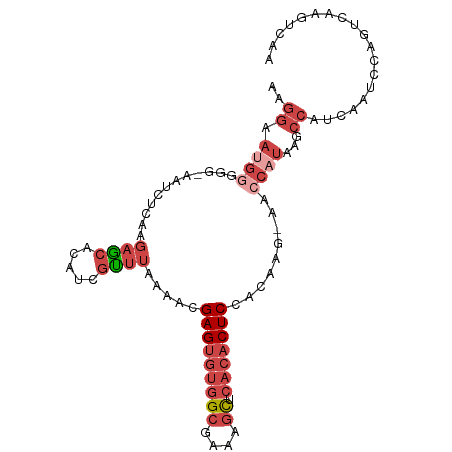
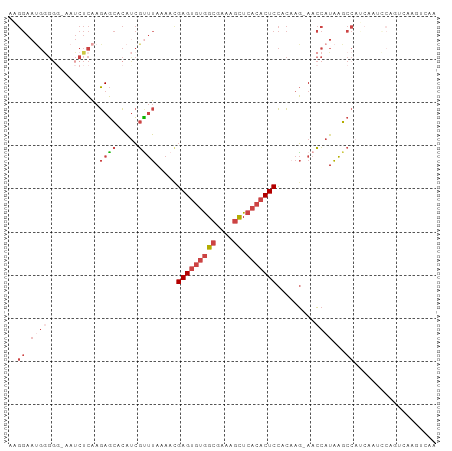
| Location | 17,068,465 – 17,068,557 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 68.72 |
| Shannon entropy | 0.60335 |
| G+C content | 0.46255 |
| Mean single sequence MFE | -21.73 |
| Consensus MFE | -12.46 |
| Energy contribution | -14.07 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.70 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.712498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17068465 92 - 21146708 UUGACUUGACUGGAUG---GGCUUAUGGUU-CUUGUGGAGUGUGAGCUUUCGCCACACUCGUUUUAAACGAUGUGCUCUUGAGAUU---CCCAUUCCUU ...........(((((---((((((.(((.-(((((.(((((((.((....))))))))).......)))).).)))..))))...---)))))))... ( -25.40, z-score = -1.45, R) >droSim1.chr2R 15717359 98 - 19596830 UUGACUUGACUGGAUGGAUGGCUUAUGGUU-CUUGUGGAGUGUGAGCUUUCGCCACACUCAUUUUAAACGAUGUGCUCUUGAGAUUCCCCCCGUUCCUU .......((.(((..(((...((((.(((.-(((((.(((((((.((....))))))))).......)))).).)))..))))..)))..))).))... ( -25.90, z-score = -1.18, R) >droSec1.super_9 394906 97 - 3197100 UUGACUUGACUGGAUGGAUGGCUUAUGGUU-CUUGUGGAGUGUGAGCUUUCGCCACACUCGUUUUAAGCGAUGUGCUCUUGAGAUU-CCCCCAUUCCUU ...........((((((..((.........-......(((((((.((....)))))))))((((((((.(.....).)))))))).-)).))))))... ( -24.80, z-score = -0.45, R) >droYak2.chr2R 9011332 83 + 21139217 --------------UUGAUGGCUUAUGGUU-CUUGUGGAGUGUGAGCUUUCGCCACACUCGUUUUAAACGAUGUGCUCUUGAGAUU-CCCCCAUUCCUC --------------(..(.(((.(((.(((-......(((((((.((....)))))))))......))).))).))).)..)....-............ ( -20.50, z-score = -1.17, R) >droEre2.scaffold_4845 11211123 88 - 22589142 ---------UUGACUUGGUGGCUUAUGGUU-CUGGUGGAGUGUGAGCUUUCGCCACACUCGUUUUAAACGGUGUGCUCUUGAGAUU-CCCUCAUUCCCU ---------....((..(.(((.(((.(((-..((..(((((((.((....)))))))))..))..))).))).))).)..))...-............ ( -24.90, z-score = -0.94, R) >droWil1.scaffold_180700 4809228 84 - 6630534 CGUACAUUUGCCACUUGUUGGCCCAUUCUUAGGUAUAGAGCUUCAACUUUCUCU---CUCAUUUUGGCACACACAGUUUCCUAAUCU------------ ........(((((.......(((........)))..((((..........))))---.......)))))..................------------ ( -8.90, z-score = 0.98, R) >consensus UUGACUUGACUGGAUGGAUGGCUUAUGGUU_CUUGUGGAGUGUGAGCUUUCGCCACACUCGUUUUAAACGAUGUGCUCUUGAGAUU_CCCCCAUUCCUU ...................(((.(((.(((.......(((((((.((....)))))))))......))).))).)))...................... (-12.46 = -14.07 + 1.61)
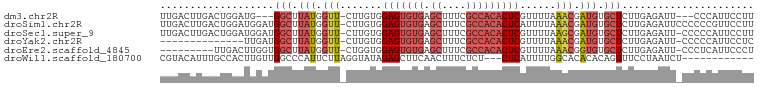

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:04 2011