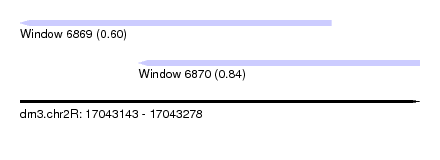
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,043,143 – 17,043,278 |
| Length | 135 |
| Max. P | 0.835776 |

| Location | 17,043,143 – 17,043,248 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 77.60 |
| Shannon entropy | 0.45106 |
| G+C content | 0.45572 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -15.66 |
| Energy contribution | -15.22 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.600036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17043143 105 - 21146708 ----------------UUUCAUUUGCAGGCAUUCUCCGUACUGUUCGUGUGCAUAGCGAUAACGUCGGAUCUGGUCUGUUUAUUCUUCAUACCCGUUCACUGGCUGCUCUUUGCAGCGAGC ----------------....((..((((((....((((.((...((((.......))))....))))))....))))))..))................((.(((((.....))))).)). ( -28.50, z-score = -1.09, R) >droSec1.super_9 369188 105 - 3197100 ----------------UUUCAUUUGCAGGCAUUCUCCGUACUGUUCGUGUGCAUAGCGAUAACGUCGGAUCUGGUCUGUUUAUUCUUCAUACCCGUUCACUGGCUGCUCUUCGCAGCGAGC ----------------....((..((((((....((((.((...((((.......))))....))))))....))))))..))................((.(((((.....))))).)). ( -27.90, z-score = -0.83, R) >droYak2.chr2R 8985446 105 + 21139217 ----------------UUUCAUUUGCAGGCAUUCUCCGUACUGUUCGUGUGCAUAGCGAUAACGUCGGAUCUGGUCUGUUUAUUCUUCAUACCCGUUCACUGGCUGCUCUUCGCAGCAAGC ----------------....((..((((((....((((.((...((((.......))))....))))))....))))))..))................((.(((((.....))))).)). ( -25.80, z-score = -0.63, R) >droEre2.scaffold_4845 11185146 105 - 22589142 ----------------UUUCAUUUGCAGGCAUUCUCCGUACUGUUCGUGUGCAUAGCGAUAACGUCGGAUCUGGUCUGUUUAUUCUUCAUACCCGUUCACUGGCUGCUCUUUGCAGCAAGC ----------------....((..((((((....((((.((...((((.......))))....))))))....))))))..))................((.(((((.....))))).)). ( -26.40, z-score = -0.81, R) >droAna3.scaffold_13266 12964767 104 + 19884421 -----------------UUAUAUUGCAGGCCUUCUCCGUGCUAUUCGUAUGCAUAGCGAUAACGUCGGAUCUGGUCUGUUUGUUCUUCAUACCCGUUCACUGGCUGCUCUUCGCGGCCAGC -----------------.......(((((((...((((......((((.......))))......))))...)))))))....................((((((((.....)))))))). ( -34.30, z-score = -2.88, R) >dp4.chr3 17679693 105 - 19779522 ---------------UUACUUG-UACAGGCAUUCUCCGUGCUAUUCGUCUGUAUUGCGAUAACAUCGGAUCUGGUCUGCUUAUUUUUCAUACCCGUUCACUGGCUGCUCUUUGCGGCCAGC ---------------.......-..(((((....((((((.(((.(((.......)))))).)).))))....))))).....................((((((((.....)))))))). ( -29.80, z-score = -2.44, R) >droWil1.scaffold_180699 1844401 105 - 2593675 ----------------UCCUUUUUGCAGGCAUUCUCCGUACUUUUCGUGUGCAUAGCGAUUACAUCGGAUUUGGUGUGUCUAUUUUUCAUACCAGUUCACUGGCUACUCUUUUUGGCCAGC ----------------......((((.((......))((((.......))))...))))........((.((((((((.........)))))))).)).(((((((.......))))))). ( -27.20, z-score = -2.39, R) >droMoj3.scaffold_6496 16019650 111 - 26866924 ---------AUACAUU-UUGUGCUCCAGGCAUUUUCUGUGCUAUUCGUGUGCAUUGCAAUAACAUCGGAUCUGGUGUGCUUGUUCUUCAUACCCGUACAUUGGCUGCUCUUUGCGGCCAGC ---------.......-.(((((....(((((.....)))))....(((((....((((..((((((....))))))..))))....)))))..)))))((((((((.....)))))))). ( -35.40, z-score = -2.42, R) >droGri2.scaffold_15245 14304213 120 + 18325388 AUACAUUUUACCCACU-UUGUGCAACAGGCCUUUUCCGUGCUAUUUGUGUGCAUUGCAAUAACAUCGGAUCUGGUGUGUUUGUUCUUCAUACCCGUACAUUGGCUGCUCUUUGCGGCCAGC ................-.(((((((((.(((...((((((.((((.(((.....))))))).)).))))...))).))))((.....)).....)))))((((((((.....)))))))). ( -33.60, z-score = -1.57, R) >droVir3.scaffold_12875 9691888 112 + 20611582 ---------UUACAUUGACUUGCUGCAGGCAUUUUCUGUGCUUUUUGUGUGCAUUGCAAUAACAUCGGAUCUGGUGUGUUUGUUCUUCAUACCCGUACAUUGGCUGCUUUUUGCAGCCAGC ---------.(((((((...((((((((((((.....)))))...)))).)))...))))............((((((.........)))))).)))..((((((((.....)))))))). ( -32.50, z-score = -1.31, R) >anoGam1.chr3L 5802778 104 - 41284009 -----------------UUCUAUUAUAGGCAUUUUCGGUGCUGUUCGUCUGCAUAGCGGUGACAUCCGAUUUGGUAUGUCUAUUUUUCAUCCCCGUUCAGUGGCUAUUUUUUGCCGCCAGU -----------------..........(((.........((((..((.((((...)))).(((((((.....)).))))).............))..))))(((........))))))... ( -25.40, z-score = -1.29, R) >apiMel3.GroupUn 386722152 105 + 399230636 ----------------UUAUAUUUUCAGGCCUUCUCUGUAUUUUUUAUUUGUAUUGCUCUUACAUCAGAUAUGAUUUGUUUCUUUUUCAUACCAGUACAUUGGUUAUUUUUUGCUGCUAGC ----------------...........(((...................(((((((.(((......)))(((((............))))).)))))))..(((........))))))... ( -9.50, z-score = 1.25, R) >consensus ________________UUUCUUUUGCAGGCAUUCUCCGUACUGUUCGUGUGCAUAGCGAUAACAUCGGAUCUGGUCUGUUUAUUCUUCAUACCCGUUCACUGGCUGCUCUUUGCAGCCAGC .........................(((......((((......((((.......))))......))))...(((.((.........)).)))......)))(((((.....))))).... (-15.66 = -15.22 + -0.45)


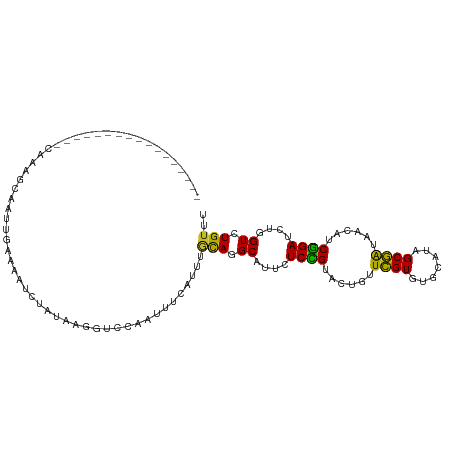
| Location | 17,043,183 – 17,043,278 |
|---|---|
| Length | 95 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 64.77 |
| Shannon entropy | 0.73173 |
| G+C content | 0.40687 |
| Mean single sequence MFE | -19.96 |
| Consensus MFE | -11.64 |
| Energy contribution | -10.84 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.46 |
| Mean z-score | -0.32 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.835776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17043183 95 - 21146708 ----------------CCAAAGUGAUUGUAACUCUAUACGGUCCAAUUUCAUUUGCAGGCAUUCUCCGUACUGUUCGUGUGCAUAGCGAUAACGUCGGAUCUGGUCUGUUU ----------------.....(.(((((((......))))))))..........((((((....((((.((...((((.......))))....))))))....)))))).. ( -22.50, z-score = -0.81, R) >droSec1.super_9 369228 95 - 3197100 ----------------CCAAAGUGAUUGUAACUCUAUACGGUCCAAUUUCAUUUGCAGGCAUUCUCCGUACUGUUCGUGUGCAUAGCGAUAACGUCGGAUCUGGUCUGUUU ----------------.....(.(((((((......))))))))..........((((((....((((.((...((((.......))))....))))))....)))))).. ( -22.50, z-score = -0.81, R) >droYak2.chr2R 8985486 99 + 21139217 ------------GCAGGCAAAGCAAUUGAAACUCUAUACGGUACAAUUUCAUUUGCAGGCAUUCUCCGUACUGUUCGUGUGCAUAGCGAUAACGUCGGAUCUGGUCUGUUU ------------((((((...((((.(((((...............))))).))))........((((.((...((((.......))))....))))))....)))))).. ( -22.56, z-score = -0.02, R) >droEre2.scaffold_4845 11185186 99 - 22589142 ------------CGAGCCAAAGCGAUUGAAACUCUAUACGGUCCAAUUUCAUUUGCAGGCAUUCUCCGUACUGUUCGUGUGCAUAGCGAUAACGUCGGAUCUGGUCUGUUU ------------.((((....((((.(((((...............))))).))))((((....((((.((...((((.......))))....))))))....)))))))) ( -19.76, z-score = 1.20, R) >droAna3.scaffold_13266 12964807 90 + 19884421 --------------------UCAAAACCAAUACUUAUCAUCACCAAUUAUAU-UGCAGGCCUUCUCCGUGCUAUUCGUAUGCAUAGCGAUAACGUCGGAUCUGGUCUGUUU --------------------................................-.(((((((...((((......((((.......))))......))))...))))))).. ( -19.80, z-score = -1.97, R) >dp4.chr3 17679733 82 - 19779522 --------------------------UUUAA---UAUAUGUUCUGAUUACUUGUACAGGCAUUCUCCGUGCUAUUCGUCUGUAUUGCGAUAACAUCGGAUCUGGUCUGCUU --------------------------.....---.....................(((((....((((((.(((.(((.......)))))).)).))))....)))))... ( -14.70, z-score = -0.18, R) >droWil1.scaffold_180699 1844441 92 - 2593675 -------------------UAACAAUGAUAAAAACCAAACACGAAAUCCUUUUUGCAGGCAUUCUCCGUACUUUUCGUGUGCAUAGCGAUUACAUCGGAUUUGGUGUGUCU -------------------.......((((...((((((..(((((((((...((((.((................)).)))).)).))))...)))..)))))).)))). ( -22.49, z-score = -1.96, R) >droGri2.scaffold_15245 14304253 111 + 18325388 UCAUAAUUGUUAUAUAUACAUUUUAUUAAUUAUACAUUUUACCCACUUUGUGCAACAGGCCUUUUCCGUGCUAUUUGUGUGCAUUGCAAUAACAUCGGAUCUGGUGUGUUU ..(((((((.((...........)).)))))))....................((((.(((...((((((.((((.(((.....))))))).)).))))...))).)))). ( -17.70, z-score = 0.57, R) >droVir3.scaffold_12875 9691928 100 + 20611582 -------UGCAAAAC-UUUAUAAUAUUAAUUUGGUUUUACAUUGACU---UGCUGCAGGCAUUUUCUGUGCUUUUUGUGUGCAUUGCAAUAACAUCGGAUCUGGUGUGUUU -------.(.(((((-(..((.......))..)))))).)((((...---((((((((((((.....)))))...)))).)))...)))).((((((....)))))).... ( -18.60, z-score = 0.63, R) >anoGam1.chr3L 5802818 98 - 41284009 -----------UGACUUAUGUGCCGUUUCUUAU-UUUUACGUCCACUUC-UAUUAUAGGCAUUUUCGGUGCUGUUCGUCUGCAUAGCGGUGACAUCCGAUUUGGUAUGUCU -----------.(((....((((((..((.((.-...)).(((.(((.(-(((....(((((.....)))))((......)))))).))))))....))..))))))))). ( -19.00, z-score = 0.14, R) >consensus _________________CAAAGCAAUUGAAAAUCUAUAAGGUCCAAUUUCAUUUGCAGGCAUUCUCCGUACUGUUCGUGUGCAUAGCGAUAACAUCGGAUCUGGUCUGUUU ......................................................(((.((....((((......((((.......))))......))))....)).))).. (-11.64 = -10.84 + -0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:40:01 2011