| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,042,966 – 17,043,081 |
| Length | 115 |
| Max. P | 0.958127 |

| Location | 17,042,966 – 17,043,081 |
|---|---|
| Length | 115 |
| Sequences | 11 |
| Columns | 122 |
| Reading direction | reverse |
| Mean pairwise identity | 69.39 |
| Shannon entropy | 0.66950 |
| G+C content | 0.39143 |
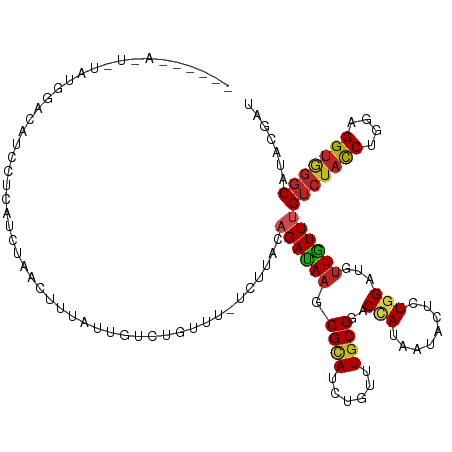
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -14.81 |
| Energy contribution | -14.71 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.958127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
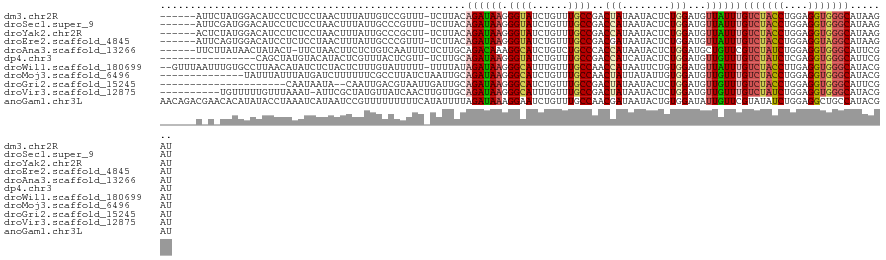
>dm3.chr2R 17042966 115 - 21146708 ------AUUCUAUGGACAUCCUCUCCUAACUUUAUUGUCCGUUU-UCUUACAGAUAAGGGUAUCUGUUUGCCGACUAUAAUACUCUGGAUGUUAUUUGUCUACCUGGAGGUGGGCAUAAGAU ------..(((...(((((((..........((((.(((.((..-....(((((((....)))))))..)).))).))))......)))))))...((((((((....))))))))..))). ( -30.99, z-score = -1.76, R) >droSec1.super_9 369011 115 - 3197100 ------AUUCGAUGGACAUCCUCUCCUAACUUUAUUGCCCGUUU-UCUUACAGAUAAGGGUAUCUGUUUGCCGACCAUAAUACUCUGGAUGUUAUUUGUCUACCUGGAGGUGGGCAUAAGAU ------....((.((....))))......(((...((((((..(-((....(((((((((((......))))..(((........)))......)))))))....)))..)))))).))).. ( -29.30, z-score = -0.84, R) >droYak2.chr2R 8985273 115 + 21139217 ------ACUCUAUGGACAUCCUCUCCUAACUUUAUUGCCCGCUU-UCUUACAGAUAAGGGUAUCUGUUUGCCGACCAUAAUACUCUGGAUGUUAUUUGUCUACCUGGAGGUGGGCAUAAGAU ------.......(((.......)))...(((...(((((((((-((....(((((((((((......))))..(((........)))......)))))))....))))))))))).))).. ( -32.60, z-score = -1.97, R) >droEre2.scaffold_4845 11184973 115 - 22589142 ------AUUCAGUGGACAUCCUCUCCUAACUUUAUUGCCCGUUU-UCUUACAGAUAAGGGUAUCUGUUUGCCGACGAUAAUACUCUGGAUGUUAUUUGUCUACCUGGAGGUAGGCAUAAGAU ------........(((((((...........(((((..((((.-.(..(((((((....)))))))..)..)))).)))))....)))))))...((((((((....))))))))...... ( -29.86, z-score = -1.11, R) >droAna3.scaffold_13266 12964598 115 + 19884421 ------UUCUUAUAACUAUACU-UUCUAACUUCUCUGUCAAUUUCUCUUGCAGACAAAGGCAUCUGUCUGCCCACCAUAAUACUCUGGAUGCUGUUCGUCUAUCUGGAGGUGGGCAUUCGAU ------................-..........((((.(((......)))))))...((((....))))(((((((.......((..((((.........))))..)))))))))....... ( -26.11, z-score = -1.66, R) >dp4.chr3 17679514 105 - 19779522 ----------------CAGCUAUGUACAUACUCGUUUACUCGUU-UCUUGCAGAUAAGGGUAUCUGUUUGCCGACCAUCAUACUCUGGAUGUUGUUUGUCUAUCUCGAGGUGGGCAUUCGAU ----------------.......................(((..-.(..(((((((....)))))))..).)))(((........)))..((((..((((((((....))))))))..)))) ( -21.80, z-score = 0.84, R) >droWil1.scaffold_180699 1844218 119 - 2593675 --GUUUAAUUUGUGCCUUAACAUAUCUCUACUCUUUGUAUUUUU-UUUUAUAGAUAAGGGCAUUUGUUUGCCAACCAUAAUUCUGUGGAUGUUAUUUGUCUACCUUGAGGUGGGCAUACGAU --........(((((((......((((((((.....))).....-.....((((((((((((......))))..(((((....)))))......))))))))....)))))))))))).... ( -28.70, z-score = -2.26, R) >droMoj3.scaffold_6496 16019465 108 - 26866924 --------------UAUUUAUUUAUGAUCUUUUUUCGCCUUAUCUAAUUGCAGAUAAGGGCAUCUGUUUGCCAACUAUUAUAUUGUGGAUGUUGUUUGUCUACCUGGAGGUGGGCAUACGAU --------------.((((((.((((((..........(((((((......)))))))((((......))))....))))))..))))))(((((.((((((((....)))))))).))))) ( -31.90, z-score = -3.24, R) >droGri2.scaffold_15245 14304050 99 + 18325388 ---------------------CAAUAAUA--CAAUUGACGUAAUUGAUUGCAGAUAAGGGCAUCUGUUUGCCGACUAUAAUACUCUGGAUGUUGUUUGUCUACCUGGAGGUGGGCAUUCGAU ---------------------........--((((..(.....)..))))........((((......))))..................((((..((((((((....))))))))..)))) ( -23.20, z-score = -0.56, R) >droVir3.scaffold_12875 9691724 111 + 20611582 ----------UGUUUUUGUUUAAAU-AUUCGCUAUGUUAUCAACUUGUUGCAGAUAAGGGCAUUUGUUUGCCGACUAUAAUACUCUGGAUGUUGUUUGUCUAUCUGGAGGUGGGCAUACGAU ----------............(((-(((((...((((((...((((((...))))))((((......))))....))))))...))))))))((.((((((((....)))))))).))... ( -25.70, z-score = -0.88, R) >anoGam1.chr3L 5802587 122 - 41284009 AACAGACGAACACAUAUACCUAAAUCAUAAUCCGUUUUUUUUUCAUAUUUUAGAUAAAGGAAUCUGUUUGCCAACGAUAAUACUGUGGAUAUUGUUCGUAUAUCUGGAGGCUGCCAUACGAU ..((((((((((.((((.((.............((((((((.((........)).)))))))).((((....))))..........))))))))))))....))))................ ( -16.00, z-score = 1.57, R) >consensus ______A_U_UAUGGACAUCCUCAUCUAACUUUAUUGUCUGUUU_UCUUACAGAUAAGGGCAUCUGUUUGCCGACCAUAAUACUCUGGAUGUUGUUUGUCUACCUGGAGGUGGGCAUACGAU ...................................................((((((.((((......))))..(((........)))...))))))(((((((....)))))))....... (-14.81 = -14.71 + -0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:59 2011