| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,031,936 – 17,032,027 |
| Length | 91 |
| Max. P | 0.978326 |

| Location | 17,031,936 – 17,032,027 |
|---|---|
| Length | 91 |
| Sequences | 13 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 68.17 |
| Shannon entropy | 0.64887 |
| G+C content | 0.56454 |
| Mean single sequence MFE | -29.71 |
| Consensus MFE | -15.68 |
| Energy contribution | -15.47 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.74 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
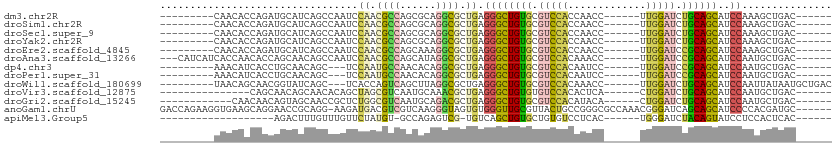
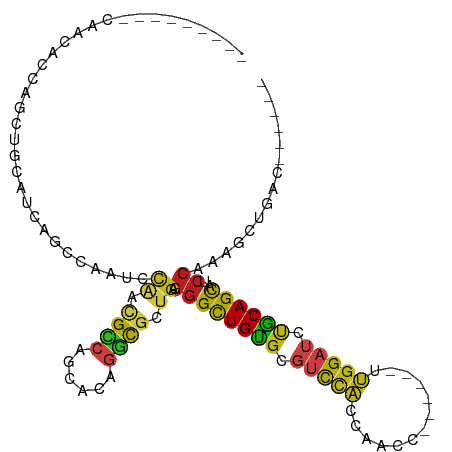
>dm3.chr2R 17031936 91 + 21146708 ---------CAACACCAGAUGCAUCAGCCAAUCCAACGCCAGCGCAGGCGCUGAGGGCUGUGCGUCCACCAACC------UUGGAUCUGCAGCAUCCAAAGCUGAC------ ---------........((((((.(((((....((.((((......)))).))..)))))))))))..(((...------.))).....((((.......))))..------ ( -31.50, z-score = -1.59, R) >droSim1.chr2R 15689443 91 + 19596830 ---------CAACACCAGAUGCAUCAGCCAAUCCAACGCCAGCGCAGGCGCUGAGGGCUGUGCGUCCACCAACC------UUGGAUCUGCAGCAUCCAAAGCUGAC------ ---------........((((((.(((((....((.((((......)))).))..)))))))))))..(((...------.))).....((((.......))))..------ ( -31.50, z-score = -1.59, R) >droSec1.super_9 360612 91 + 3197100 ---------CAACACCAGAUGCAUCAGCCAAUCCAACGCCAGCGCAGGCGCUGAGGGCUGUGCGUCCACCAACC------UUGGAUCUGCAGCAUCCAAAGCUGAC------ ---------........((((((.(((((....((.((((......)))).))..)))))))))))..(((...------.))).....((((.......))))..------ ( -31.50, z-score = -1.59, R) >droYak2.chr2R 8976863 91 - 21139217 ---------CAACACCAGAUGCAUCAGCCAAUCCAACGCCAGCGCAGGCGCUGAGGGCUGUGCGUCCACCAACC------UUGGAUCUGCAGCAUCCAAAGCUGAC------ ---------........((((((.(((((....((.((((......)))).))..)))))))))))..(((...------.))).....((((.......))))..------ ( -31.50, z-score = -1.59, R) >droEre2.scaffold_4845 11176491 91 + 22589142 ---------CAACACCAGAUGCAUCAGCCAAUCCAACGCCAGCAAAGGCGCUGAGGGCUGUGCGUCCACCAACC------UUGGAUCCGCAGCAUCCAAAGCUGAC------ ---------........((((((.(((((....((.((((......)))).))..)))))))))))..(((...------.))).....((((.......))))..------ ( -31.50, z-score = -2.48, R) >droAna3.scaffold_13266 12956279 97 - 19884421 ---CAUCAUCACCAACACCAGCAACAGCCAAUCCAACGCCAGCAUAGGCGCUGAGGGCUGUGCGUCCACAAACC------UUGGAUCCGCAGCAUCCAAUGCUGAC------ ---...............(((((((((((....((.((((......)))).))..))))))(((((((......------.))))..))).........)))))..------ ( -31.30, z-score = -2.52, R) >dp4.chr3 17670200 88 + 19779522 ---------AAACAUCACCUGCAACAGC---UCCAAUGCCAACACAGGCGCUGAGGGCUGUGCGUCCACAAUCC------UUGGAUCCGCAGCAUCCAAUGCUGAC------ ---------...........(((.((((---((((.((((......)))).)).)))))))))(((((......------.)))))...((((((...))))))..------ ( -30.20, z-score = -2.35, R) >droPer1.super_31 815241 88 - 935084 ---------AAACAUCACCUGCAACAGC---UCCAAUGCCAACACAGGCGCUGAGGGCUGUGCGUCCACAAUCC------UUGGAUCCGCAGCAUCCAAUGCUGAC------ ---------...........(((.((((---((((.((((......)))).)).)))))))))(((((......------.)))))...((((((...))))))..------ ( -30.20, z-score = -2.35, R) >droWil1.scaffold_180699 1834902 94 + 2593675 ---------UAACAGCAACGGUAUCAGC---UCACCAGUCAGCUUAGGCGCUGAGGGCUGUGCGUCCACAAACC------UUGGAUCUGCAGCAUCCAAUUAUAAUGCUGAC ---------...(((((..(((......---..)))..(((((......)))))((((((((.(((((......------.))))).))))))..))........))))).. ( -29.60, z-score = -1.47, R) >droVir3.scaffold_12875 9682733 85 - 20611582 ---------------CAGCAACAGCAACACAGCUAGCGUCAAUGCAAACGCUGAGGGCUGUGUGUCCACACUCA------CUGGAUCUGCAGCAUCCAAUGCUGAC------ ---------------......((((((((((((((((((........)))))...))))))))...........------.((((((....).))))).)))))..------ ( -28.50, z-score = -1.04, R) >droGri2.scaffold_15245 14294944 88 - 18325388 ------------CAACAACAGUAGCAACCGCUCUGGCGUCAAUGCAGACGCUGAGGGCUGUGCGUCCACAUACA------CUGGAUCUGCAGCAUCCAAUGCUGAC------ ------------......(((..(((...(((((((((((......)))))).)))))..)))(((((......------.))))))))((((((...))))))..------ ( -30.90, z-score = -1.52, R) >anoGam1.chrU 8539713 105 + 59568033 GACCAGAAGGUGAAGCAGGAACCGCAGG-AAGAUGACGUCGUCAAGGGUAGUGUGGGUUGCGUUACUGCCGGGCGCCAAACGGGAUCAGCAGCAUCCCCACGAUGC------ .(((....(((.........))).....-..((((....))))...))).(((.(((.((((((..(.(((.........))).)..))).))).)))))).....------ ( -27.90, z-score = 1.73, R) >apiMel3.Group5 2665599 79 - 13386189 -------------------AGACUUUGUUUGUUCUAUGU-GCCAGAGUCG-UGUCAGCUGUGCUGUGUCCUCAC------UGGGAUCUACAGUAUCCUCCACUCAC------ -------------------.(((((((...((.......-)))))))))(-((..((..((((((((((((...------.)))))..))))))).)).)))....------ ( -20.10, z-score = -0.26, R) >consensus _________CAACACCAGCUGCAUCAGCCAAUCCAACGCCAGCACAGGCGCUGAGGGCUGUGCGUCCACCAACC______UUGGAUCUGCAGCAUCCAAAGCUGAC______ .................................((.((((......)))).)).((((((((.(((((.............))))).))))))..))............... (-15.68 = -15.47 + -0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:58 2011