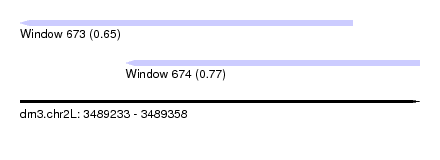
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,489,233 – 3,489,358 |
| Length | 125 |
| Max. P | 0.773446 |

| Location | 3,489,233 – 3,489,337 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 68.08 |
| Shannon entropy | 0.66550 |
| G+C content | 0.31973 |
| Mean single sequence MFE | -14.27 |
| Consensus MFE | -4.86 |
| Energy contribution | -4.62 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.652367 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 3489233 104 - 23011544 --AAAAUCGC-AAAUCUCGCACUAUAUUCCAUCAUC----GAAUUUGC--AUAUGCAAAUUUCAUUCCCACGACUUUUAAUUGCAACGAACACACAAAAACUAAAACUGGACA----- --......((-.......)).......((((.....----((((((((--....))))))))........((..............))...................))))..----- ( -9.54, z-score = -0.37, R) >droSim1.chr2L 3442578 104 - 22036055 --AAAAUCGC-AAAUCUCGCACUAUAUUCCAUCAUC----GAAUUUGC--AUAUGCAAAUUUCAUUCCCACGACUUUUAAUUGCAACGAACACACAAAAACUAAAACUGGACA----- --......((-.......)).......((((.....----((((((((--....))))))))........((..............))...................))))..----- ( -9.54, z-score = -0.37, R) >droSec1.super_129 51228 105 - 51580 --AAAAUCGCAAAAUCUCGCACUAUAUUCCAUCAUC----GAAUUUGC--AUAUGCAAAUUUCAUUCCCACGACUUUUAAUUGCAACGAACACACAAAAACUAAAACUGGACA----- --......((........)).......((((.....----((((((((--....))))))))........((..............))...................))))..----- ( -9.84, z-score = -0.53, R) >droYak2.chr2L 3477177 102 - 22324452 --AAAAUCGC-AAAUCUCGCACUAUAUUCCAUCAUC----GAAUUUGC--AUAUGCAAAUUUCAUUCCCACGACUUUUAAUUGCGAUGA--AAACAGAAACUAAAACUGGACA----- --...(((((-((.......................----((((((((--....))))))))...((....)).......)))))))..--...(((.........)))....----- ( -14.10, z-score = -1.02, R) >droEre2.scaffold_4929 3524011 102 - 26641161 --AAAAUCGC-AAAUCUCGCACUAUAUUCCAUCAUC----GAAUUUGC--AUAUGCAAAUUUCAUUCCCACGACUUUUAAUUGCAAUGA--AAACAAAAACUAAAACUGGACA----- --......((-.......)).......((((.....----...(((((--....)))))(((((((..................)))))--))..............))))..----- ( -10.37, z-score = -0.37, R) >droAna3.scaffold_12916 5535774 90 - 16180835 --AAAUUCGC-AAAUCCCAAAUAGUAUUCCAUCAUC----AGAUUUGC--AUAUGCAAAUUUCAUUCCCACGACUUUUAAUUGCAA-GACUAGAGAAGAA------------------ --...(((((-(((((....................----.)))))))--...(((((.......((....)).......))))).-.......)))...------------------ ( -10.24, z-score = -0.53, R) >dp4.chr4_group3 6432661 91 + 11692001 --ACAUUCGCAAAAUCUUUAACCGUAUUCCAUCAUCAGAUGGAUUUGC--AUAUGUAAAUUUCAUUCCCACGAUUUUUAAUUGAAA-------ACAAAAGCU---------------- --......((((((((...........((((((....))))))(((((--....)))))............))))))...(((...-------.)))..)).---------------- ( -13.70, z-score = -1.60, R) >droPer1.super_1 3532802 91 + 10282868 --AAAUUCGCAAAAUCUUUAACUGUAUUCCAUCAUCAGAUGGAUUUGC--AUAUGCAAAUUUCAUUCCCACGAUUUUUAAUUGAAA-------ACAAAAGCU---------------- --......((((((((...........((((((....))))))(((((--....)))))............))))))...(((...-------.)))..)).---------------- ( -15.70, z-score = -1.88, R) >droWil1.scaffold_180708 8954612 95 - 12563649 --AAAUUCGCAACAA---AAAAAAAAUUCGAUCA------CAAUUUGC--AUAUGCAAAUUGUAUUCCCACGACUUUUAAUUUUUUUUCGCAAAAAAAAAUCCAAAUA---------- --......((...((---((((((((.(((...(------((((((((--....))))))))).......))).))))..))))))...)).................---------- ( -13.80, z-score = -2.43, R) >droVir3.scaffold_12963 4910358 105 + 20206255 --AAAUUCGCCAAAA---AAAAAAAAGACGAAUAUC----UGAUUUGC--AUAUGCAAAUUUGAUUCCCACGACUUUAAAUUGUGUGCGAAAAACACAAAAUCAAACAAAAGGCCA-- --......(((....---.....((((.((...(((----.(((((((--....))))))).))).....)).))))...((((((.......))))))............)))..-- ( -21.70, z-score = -3.27, R) >droMoj3.scaffold_6500 14077263 112 + 32352404 AAAGAUUGGCUAAGAUUCACACAAUAUGCGAAUAUU----CGAUUUGCAUAUAUGCAAAUUUAAUUCCCACGACUUCAAAUUGUGUA--AAUUAAAUAAAGUCAUAUGAAAGGCCAGU ....(((((((....((((....((((((((((...----..))))))))))......((((((((..(((((.......)))))..--)))))))).........)))).))))))) ( -26.80, z-score = -3.42, R) >droGri2.scaffold_15126 4118128 101 - 8399593 --AAAUUCGCUAAA----AUAAAAGCAACAUGUAUU----CGAUUUGC--AUAUGCAAAUUCGAUUCCCACGACUUUUAAUUGUGUAC-AAAAAACCA----CAAAAAAAGCCCCAGU --......(((...----.((((((..........(----((((((((--....))))).)))).........)))))).(((((...-.......))----)))....)))...... ( -15.91, z-score = -2.30, R) >consensus __AAAUUCGC_AAAUCUCACACUAUAUUCCAUCAUC____GAAUUUGC__AUAUGCAAAUUUCAUUCCCACGACUUUUAAUUGCAA_GAA_AAACAAAAACUAAAACUGGACA_____ ........................................((((((((......))))))))........................................................ ( -4.86 = -4.62 + -0.24)
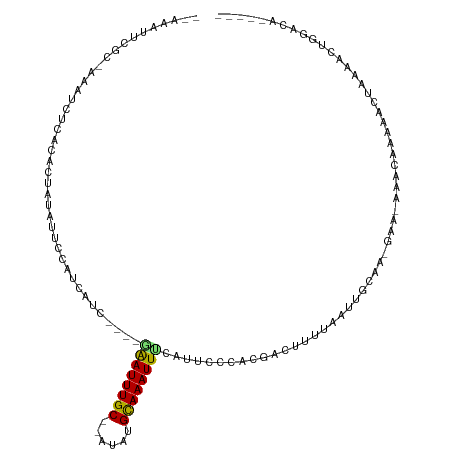

| Location | 3,489,266 – 3,489,358 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 69.23 |
| Shannon entropy | 0.64350 |
| G+C content | 0.32404 |
| Mean single sequence MFE | -12.69 |
| Consensus MFE | -4.86 |
| Energy contribution | -4.62 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.773446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3489266 92 - 23011544 -------AAUGCAAAGCA-GACACAU-----AAAAAAAUCGC-AAAUCUCGCACUAUAUUCCAUCAUC----GAAUUUGC--AUAUGCAAAUUUCAUUCCCACGACUUUUAA -------..(((...)))-.......-----..((((.((((-.......))................----((((((((--....)))))))).........)).)))).. ( -9.40, z-score = -0.47, R) >droSim1.chr2L 3442611 92 - 22036055 -------AAUGCAAAGCA-GGCACAU-----AAAAAAAUCGC-AAAUCUCGCACUAUAUUCCAUCAUC----GAAUUUGC--AUAUGCAAAUUUCAUUCCCACGACUUUUAA -------..(((......-.)))...-----..((((.((((-.......))................----((((((((--....)))))))).........)).)))).. ( -10.30, z-score = -0.11, R) >droSec1.super_129 51261 93 - 51580 -------AAUGCAAAGCA-GACACAU-----AAAAAAAUCGCAAAAUCUCGCACUAUAUUCCAUCAUC----GAAUUUGC--AUAUGCAAAUUUCAUUCCCACGACUUUUAA -------..(((...)))-.......-----..((((.((((........))................----((((((((--....)))))))).........)).)))).. ( -9.70, z-score = -0.58, R) >droYak2.chr2L 3477208 93 - 22324452 -------UAUGCAAAGCA-GACACAU----AAAAAAAAUCGC-AAAUCUCGCACUAUAUUCCAUCAUC----GAAUUUGC--AUAUGCAAAUUUCAUUCCCACGACUUUUAA -------..(((...)))-.......----...((((.((((-.......))................----((((((((--....)))))))).........)).)))).. ( -9.40, z-score = -0.50, R) >droEre2.scaffold_4929 3524042 96 - 26641161 -------AAUGCAAAGCA-GACACAC-ACAUAAGAAAAUCGC-AAAUCUCGCACUAUAUUCCAUCAUC----GAAUUUGC--AUAUGCAAAUUUCAUUCCCACGACUUUUAA -------..(((...)))-.......-......((((.((((-.......))................----((((((((--....)))))))).........)).)))).. ( -9.80, z-score = -0.38, R) >droAna3.scaffold_12916 5535793 93 - 16180835 -------AAAGCAAAGCA-ACUGCCA----CAAAAAAUUCGC-AAAUCCCAAAUAGUAUUCCAUCAUC----AGAUUUGC--AUAUGCAAAUUUCAUUCCCACGACUUUUAA -------...(((..((.-...))..----..........((-(((((....................----.)))))))--...)))........................ ( -10.50, z-score = -1.33, R) >dp4.chr4_group3 6432676 103 + 11692001 -------AAACCAAAGCACAACCCACAAUGAAAAACAUUCGCAAAAUCUUUAACCGUAUUCCAUCAUCAGAUGGAUUUGC--AUAUGUAAAUUUCAUUCCCACGAUUUUUAA -------...................(((((((.((((..(((((..............((((((....)))))))))))--..))))...))))))).............. ( -16.23, z-score = -2.71, R) >droPer1.super_1 3532817 103 + 10282868 -------AAACCGAAGCACAACCCACAAUGAAAAAAAUUCGCAAAAUCUUUAACUGUAUUCCAUCAUCAGAUGGAUUUGC--AUAUGCAAAUUUCAUUCCCACGAUUUUUAA -------...................(((((((..........................((((((....))))))(((((--....)))))))))))).............. ( -15.60, z-score = -1.71, R) >droWil1.scaffold_180708 8954640 76 - 12563649 -------------------------UGCGAAAAAAAAUUCGC---AACAAAAAAAAAAUUCGAUCA------CAAUUUGC--AUAUGCAAAUUGUAUUCCCACGACUUUUAA -------------------------((((((......)))))---)...............((..(------((((((((--....)))))))))..))............. ( -18.00, z-score = -4.21, R) >droVir3.scaffold_12963 4910394 90 + 20206255 ------------UAGUUAAAUAAAAC-UUGACUAAAAUUCGCCAAAA---AAAAAAAAGACGAAUAUC----UGAUUUGC--AUAUGCAAAUUUGAUUCCCACGACUUUAAA ------------(((((((.......-))))))).............---.....((((.((...(((----.(((((((--....))))))).))).....)).))))... ( -14.70, z-score = -3.56, R) >droMoj3.scaffold_6500 14077299 97 + 32352404 -------AAGCAAUAUCAAAAUAGAA----GAAAAGAUUGGCUAAGAUUCACACAAUAUGCGAAUAUU----CGAUUUGCAUAUAUGCAAAUUUAAUUCCCACGACUUCAAA -------................(((----(....(..(((...(((((..((..((((((((((...----..)))))))))).))..))))).....))))..))))... ( -13.20, z-score = -0.50, R) >droGri2.scaffold_15126 4118161 102 - 8399593 AAAGCAACACAAUGCAUAAACUAGCCAACAACUAAAAUUCGCUAAAA---UAAAAGCA-ACAUGUAUU----CGAUUUGC--AUAUGCAAAUUCGAUUCCCACGACUUUUAA ...(((......))).................(((((.(((((....---....))).-........(----((((((((--....))))).)))).......)).))))). ( -15.50, z-score = -2.05, R) >consensus _______AAUGCAAAGCA_AACACAC____AAAAAAAUUCGC_AAAUCUCGAACUAUAUUCCAUCAUC____GAAUUUGC__AUAUGCAAAUUUCAUUCCCACGACUUUUAA ........................................................................((((((((......)))))))).................. ( -4.86 = -4.62 + -0.24)
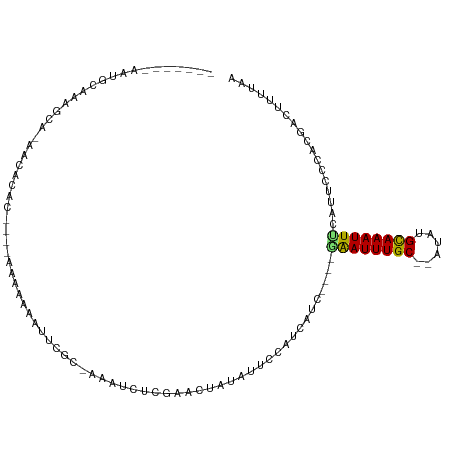
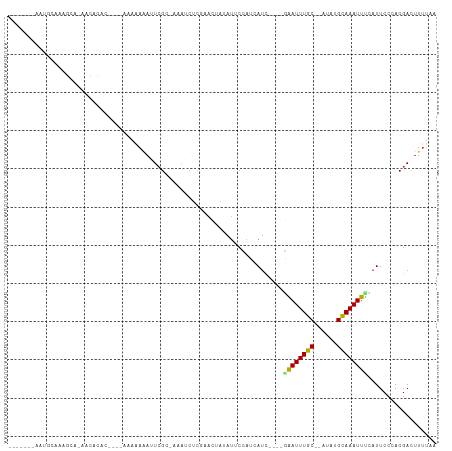
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:14:02 2011