
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,023,187 – 17,023,295 |
| Length | 108 |
| Max. P | 0.838583 |

| Location | 17,023,187 – 17,023,287 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.14 |
| Shannon entropy | 0.58297 |
| G+C content | 0.36830 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -9.52 |
| Energy contribution | -9.44 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.686844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
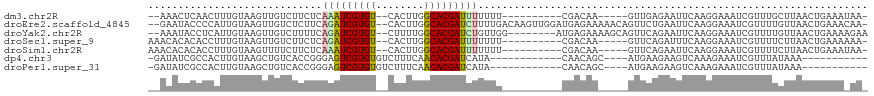
>dm3.chr2R 17023187 100 + 21146708 --AAACUCAACUUUGUAAGUUGUCUUCUCAAAUCGUGU--CACUUGGCACGAUUUUUUU----------CGACAA-----GUUGAGAAUUCAAGGAAAUCGUUUGCUUAACUGAAAUAA- --...(((((((........((((.....(((((((((--(....))))))))))....----------.)))))-----)))))).....(((.((.....)).)))...........- ( -23.50, z-score = -1.86, R) >droEre2.scaffold_4845 11167446 115 + 22589142 --GAAUACCCCAUUGUAAGUUGUCUCUUCAGAUCGUGU--CACUUGGCACGAUCUUUUGACAAGUUGGAUGAGAAAAACAGUUCUGAAUUCAAGGAAAUCGUUUUGUUAACUGAAACAA- --.......((........(((((.....(((((((((--(....))))))))))...))))).((((((.((((......))))..)))))))).....(((((.......)))))..- ( -29.20, z-score = -1.45, R) >droYak2.chr2R 8967873 108 - 21139217 --AAAUACCUCAUUGUAAGUUGUCUUUUCAGAUCGUGU--CUUUUGGCACGAUCUGUUGG--------AUGAGAAAAGCAGUUCAGAAUUCAAGGAAAUCGUUUUGUUAACUGAAAAGAA --.......((.((((...(..(((...((((((((((--(....)))))))))))..))--------)..).....))))(((((....(((((......)))))....)))))..)). ( -29.00, z-score = -2.20, R) >droSec1.super_9 351918 102 + 3197100 AAACACACACCUUUGUAAGUUGUCUUCUCAGAUCGUGU--CACUUGGCACGAUUUUUUU----------CGACAA-----GUUCAGAUUUCAAGGAAAUCGUUUUCUUAACUGAAAAAA- ...........((((.((.(((((.....(((((((((--(....))))))))))....----------.)))))-----.))))))(((((((((((....))))))...)))))...- ( -25.90, z-score = -2.59, R) >droSim1.chr2R 15680468 102 + 19596830 AAACACACACCUUUGUAAGUUUUCUUCUCAAAUCGUGU--CACUUGGCACGAUUUUUUU----------CGACAA-----GUUCAGAAUUCAAGGAAAUCGUUUUCUUAACUGAAAUAA- ...........((((.((.((.((.....(((((((((--(....))))))))))....----------.)).))-----.)))))).((((((((((....))))))...))))....- ( -19.10, z-score = -1.27, R) >dp4.chr3 17660077 92 + 19779522 -GAUAUCGCCACUUGUAAGCUGUCACCGGGAGUCGUGUGUCUUUCAACACGAUCAUA------------CAACAGC----AUGAAGAAGUCAAAGAAAUCGUUUAUAAA----------- -(((.((...((((.((.(((((......((.((((((........))))))))...------------..)))))----.))...))))....)).))).........----------- ( -19.60, z-score = -0.74, R) >droPer1.super_31 805116 92 - 935084 -GAUAUCGCCACUUGUAAGCUGUCACCGGGAGUCGUGUGUCUUUCAACACGAUCAUA------------CAACAGC----AUGAAGAAGUCAAAGAAAUCGUUUAUAAA----------- -(((.((...((((.((.(((((......((.((((((........))))))))...------------..)))))----.))...))))....)).))).........----------- ( -19.60, z-score = -0.74, R) >consensus __AAAUACCCCUUUGUAAGUUGUCUUCUCAAAUCGUGU__CACUUGGCACGAUCUUUUU__________CGACAA_____GUUCAGAAUUCAAGGAAAUCGUUUUCUUAACUGAAA_AA_ .............................(((((((((........)))))))))................................................................. ( -9.52 = -9.44 + -0.08)


| Location | 17,023,193 – 17,023,295 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.13 |
| Shannon entropy | 0.57459 |
| G+C content | 0.37038 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -10.79 |
| Energy contribution | -10.10 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.838583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
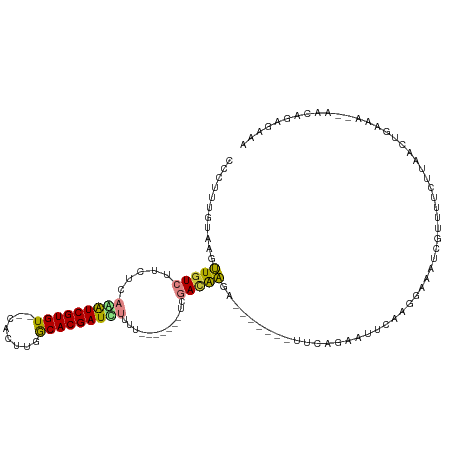
>dm3.chr2R 17023193 102 + 21146708 AACUUUGUAAGUUGUCUUCUCAAAUCGUGU--CACUUGGCACGAUUUUUU-------UCGACAAG--------UUGAGAAUUCAAGGAAAUCGUUUGCUUAACUGAAAU-AACAGAGAAA ..((((((....((((.....(((((((((--(....))))))))))...-------..))))((--------(((((.....((.(....).))..))))))).....-.))))))... ( -27.20, z-score = -2.27, R) >droEre2.scaffold_4845 11167452 117 + 22589142 CCCAUUGUAAGUUGUCUCUUCAGAUCGUGU--CACUUGGCACGAUCUUUUGACAAGUUGGAUGAGAAAAACAGUUCUGAAUUCAAGGAAAUCGUUUUGUUAACUGAAAC-AACAGAGAAA ..........(((((.((...(((((((((--(....)))))))))).((((((((((((((.((((......))))..)))))).(....)..))))))))..)).))-)))....... ( -29.90, z-score = -1.07, R) >droYak2.chr2R 8967879 110 - 21139217 CUCAUUGUAAGUUGUCUUUUCAGAUCGUGU--CUUUUGGCACGAUCUGUU--------GGAUGAGAAAAGCAGUUCAGAAUUCAAGGAAAUCGUUUUGUUAACUGAAAAGAACAGAGAAA (((.((((...(..(((...((((((((((--(....)))))))))))..--------)))..).....))))(((((....(((((......)))))....))))).......)))... ( -31.80, z-score = -2.35, R) >droSec1.super_9 351926 102 + 3197100 ACCUUUGUAAGUUGUCUUCUCAGAUCGUGU--CACUUGGCACGAUUUUUU-------UCGACAAG--------UUCAGAUUUCAAGGAAAUCGUUUUCUUAACUGAAAA-AACAGAGAAA ..((((((((.(((((.....(((((((((--(....))))))))))...-------..))))).--------))..((((((...)))))).(((((......)))))-.))))))... ( -29.10, z-score = -2.81, R) >droSim1.chr2R 15680476 102 + 19596830 ACCUUUGUAAGUUUUCUUCUCAAAUCGUGU--CACUUGGCACGAUUUUUU-------UCGACAAG--------UUCAGAAUUCAAGGAAAUCGUUUUCUUAACUGAAAU-AACAGAGAAA ..((((((....((((.....(((((((((--(....))))))))))...-------......((--------((.((((..(.........)..)))).)))))))).-.))))))... ( -23.40, z-score = -1.82, R) >dp4.chr3 17660084 85 + 19779522 CCACUUGUAAGCUGUCACCGGGAGUCGUGUGUCUUUCAACACGAUCAUAC---------AACAGCA-------UGAAGAAGUCAAAGAAAUCGUUUAUAAA------------------- ....(((((((((((......((.((((((........))))))))....---------.))))).-------(((.....)))..........)))))).------------------- ( -17.40, z-score = -0.63, R) >droPer1.super_31 805123 85 - 935084 CCACUUGUAAGCUGUCACCGGGAGUCGUGUGUCUUUCAACACGAUCAUAC---------AACAGCA-------UGAAGAAGUCAAAGAAAUCGUUUAUAAA------------------- ....(((((((((((......((.((((((........))))))))....---------.))))).-------(((.....)))..........)))))).------------------- ( -17.40, z-score = -0.63, R) >consensus CCCUUUGUAAGUUGUCUUCUCAAAUCGUGU__CACUUGGCACGAUCUUUU_______UCGACAAGA_______UUCAGAAUUCAAGGAAAUCGUUUUCUUAACUGAAA__AACAGAGAAA ....((((.............(((((((((........))))))))).............))))........................................................ (-10.79 = -10.10 + -0.69)

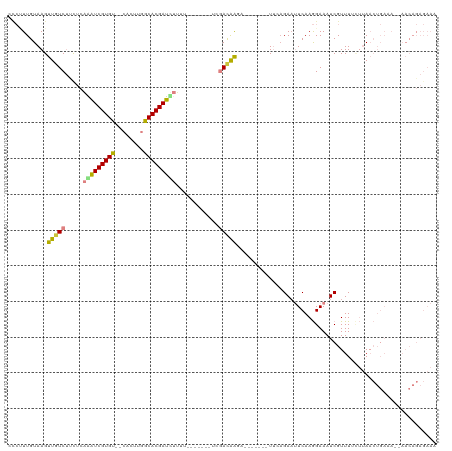
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:57 2011