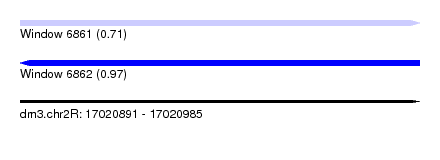
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 17,020,891 – 17,020,985 |
| Length | 94 |
| Max. P | 0.973679 |

| Location | 17,020,891 – 17,020,985 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 76.78 |
| Shannon entropy | 0.44967 |
| G+C content | 0.42644 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -15.12 |
| Energy contribution | -16.27 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.711567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
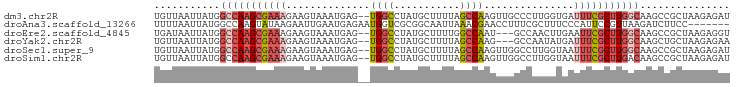
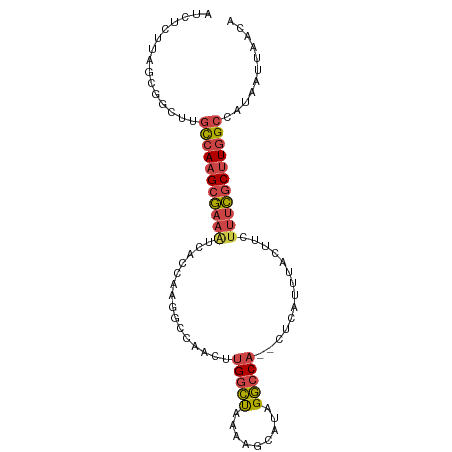
>dm3.chr2R 17020891 94 + 21146708 UGUUAAUUAUGGCCAAGCGAAAGAAGUAAAUGAG--UGGCCUAUGCUUUUAGCCAAGUUGCCCUUGGUGAUUUCGCUUGGCAAGCCGCUAAGAGAU ...........(((((((((((((((((....((--....)).))))))).((((((.....))))))...))))))))))............... ( -28.10, z-score = -1.57, R) >droAna3.scaffold_13266 12946100 89 - 19884421 UUUUAAUUAUGGCCAAGUAUAAGAAUUGAAUGAGAAUGGUCGCGGCAAUUAAACGAACCUUUCGCUUUCCCAUUCCGUUAAGAUCUUCC------- ....................((((....((((.((((((..((((................))))....))))))))))....))))..------- ( -13.09, z-score = 0.74, R) >droEre2.scaffold_4845 11165088 91 + 22589142 UGAUAAUUAUGGCCAAGCGAAAGAAGUAAAUGAG--UGGCCUAUGCUUUUGGCCAAU---GCCAACUUGAAUUCGCUUGGCAAGCCGCUAAGAGGU ...........((((((((((..((((.......--(((((.........)))))..---....))))...))))))))))..(((.......))) ( -30.12, z-score = -2.39, R) >droYak2.chr2R 8965487 91 - 21139217 UGUUAAUUAUGGCCAAGCGAAAGAAGUAAAUGAG--UGGCCUAUGCUUUUAGCCAAG---GCCAAUAUGAUUUCGCUUGGCAAGCUGCUAAGAGAA ...........(((((((((((...(((......--((((((..((.....))..))---)))).)))..)))))))))))............... ( -30.70, z-score = -2.94, R) >droSec1.super_9 349723 94 + 3197100 UGUUAAUUAUGGCCAAGCGAAAGAAGUAAAUGAG--UGGCCUAUGCUUUUAGCCAAGUUGGCCUUGGUAAUUUCGCUUGGCAAGCCGCUAAGAGAU ...........(((((((((((((((((....((--....)).))))))).((((((.....))))))...))))))))))............... ( -28.10, z-score = -1.26, R) >droSim1.chr2R 15678215 94 + 19596830 UGUUAAUUAUGGCCAAGCGAAAGAAGUAAAUGAG--UGGCCUAUGCUUUUAGCCAAGUUGGCCUUGGUAAUUUCGCUUGACAAGCCGCUAAGAGAU .(((..((((((((((((((((((((((....((--....)).))))))).((((((.....))))))...))))))))....)))).)))..))) ( -24.90, z-score = -0.93, R) >consensus UGUUAAUUAUGGCCAAGCGAAAGAAGUAAAUGAG__UGGCCUAUGCUUUUAGCCAAGUUGGCCUUGGUGAUUUCGCUUGGCAAGCCGCUAAGAGAU ...........(((((((((((..............((((...........))))...............)))))))))))............... (-15.12 = -16.27 + 1.14)
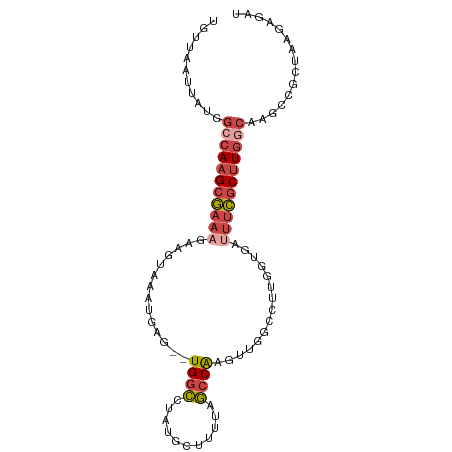
| Location | 17,020,891 – 17,020,985 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 76.78 |
| Shannon entropy | 0.44967 |
| G+C content | 0.42644 |
| Mean single sequence MFE | -23.66 |
| Consensus MFE | -16.49 |
| Energy contribution | -16.97 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 17020891 94 - 21146708 AUCUCUUAGCGGCUUGCCAAGCGAAAUCACCAAGGGCAACUUGGCUAAAAGCAUAGGCCA--CUCAUUUACUUCUUUCGCUUGGCCAUAAUUAACA ...............(((((((((((......(((....)))((((.........)))).--............)))))))))))........... ( -25.30, z-score = -2.02, R) >droAna3.scaffold_13266 12946100 89 + 19884421 -------GGAAGAUCUUAACGGAAUGGGAAAGCGAAAGGUUCGUUUAAUUGCCGCGACCAUUCUCAUUCAAUUCUUAUACUUGGCCAUAAUUAAAA -------((..((...(((..(((((((((..((...(((..........))).))....))))))))).....)))...))..)).......... ( -15.60, z-score = 0.32, R) >droEre2.scaffold_4845 11165088 91 - 22589142 ACCUCUUAGCGGCUUGCCAAGCGAAUUCAAGUUGGC---AUUGGCCAAAAGCAUAGGCCA--CUCAUUUACUUCUUUCGCUUGGCCAUAAUUAUCA ...............((((((((((...((((....---..(((((.........)))))--.......))))..))))))))))........... ( -27.12, z-score = -2.48, R) >droYak2.chr2R 8965487 91 + 21139217 UUCUCUUAGCAGCUUGCCAAGCGAAAUCAUAUUGGC---CUUGGCUAAAAGCAUAGGCCA--CUCAUUUACUUCUUUCGCUUGGCCAUAAUUAACA ...............(((((((((((......((((---((..((.....))..))))))--............)))))))))))........... ( -26.77, z-score = -3.13, R) >droSec1.super_9 349723 94 - 3197100 AUCUCUUAGCGGCUUGCCAAGCGAAAUUACCAAGGCCAACUUGGCUAAAAGCAUAGGCCA--CUCAUUUACUUCUUUCGCUUGGCCAUAAUUAACA ...............(((((((((((((((((((.....))))).)))..((....))..--............)))))))))))........... ( -24.80, z-score = -2.01, R) >droSim1.chr2R 15678215 94 - 19596830 AUCUCUUAGCGGCUUGUCAAGCGAAAUUACCAAGGCCAACUUGGCUAAAAGCAUAGGCCA--CUCAUUUACUUCUUUCGCUUGGCCAUAAUUAACA ..........(((....(((((((((((((((((.....))))).)))..((....))..--............)))))))))))).......... ( -22.40, z-score = -1.55, R) >consensus AUCUCUUAGCGGCUUGCCAAGCGAAAUCACCAAGGCCAACUUGGCUAAAAGCAUAGGCCA__CUCAUUUACUUCUUUCGCUUGGCCAUAAUUAACA ...............(((((((((((...............(((((.........)))))..............)))))))))))........... (-16.49 = -16.97 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:54 2011