
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,998,675 – 16,998,772 |
| Length | 97 |
| Max. P | 0.871561 |

| Location | 16,998,675 – 16,998,772 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 90.34 |
| Shannon entropy | 0.18407 |
| G+C content | 0.49987 |
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -25.76 |
| Energy contribution | -27.52 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871561 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
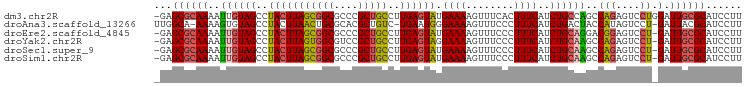
>dm3.chr2R 16998675 97 + 21146708 AAGGAUGCGCAAUCCAGGACUCUGGCUGGCAGAUGAAAGUGAAACUUUUCAUACUCAAGGCAGCGGGCGCCGCUAAGUAGGCUACAAUUUUGCGCUC- ......((((((....((.(.((.((((.(...(((..(((((....)))))..))).).)))).)).)))(((.....))).......))))))..- ( -28.60, z-score = -0.11, R) >droAna3.scaffold_13266 12922715 95 - 19884421 AAGGAUGCGUAAUC-AGGACUAUGGUAGUCAGAUGAAAGGGAAACUUUUCCCAUUCA-GACAGCGUGCGCAGUUAAGUAGGCUACACUUUU-UGCCAA ...(((.....)))-.......((((((((...((((.(((((....))))).))))-)))(((.(((........))).)))........-))))). ( -25.20, z-score = -1.09, R) >droEre2.scaffold_4845 11142911 96 + 22589142 AAGGAUGCGCAAUC-AGGACUCCGUCCUGCAGAUGAAAGGGAAACUUUUCAUACUCAAGGCAGCGGGCGCCGCUAAGUAGGCUACAAUUUUGCGCUC- ......((((((..-.((..((((((((..((((((((((....)))))))).))..)))..)))))..))(((.....))).......))))))..- ( -34.50, z-score = -2.33, R) >droYak2.chr2R 8943104 96 - 21139217 AAGGAUGCGCAAUC-AGGACUCUGGCUUGCAGAUGAAAGGGAAACUUUUCCUACUCAAGGCAGCGGACGCCACUAAGUAGGCUACAAUUUUGCGCUC- ......((((((..-.(..(.(((.((((.((..((((((....))))))...)))))).))).)..)(((........))).......))))))..- ( -34.20, z-score = -2.91, R) >droSec1.super_9 328099 96 + 3197100 AAGGAUGCGCAAUC-AGGACUCUGGCUUGCAGAUGAAAGGGAAACUUUUCAUACUCAAGGCAGCGGGCGCCGCUAAGUAGGCUACAAUUUUGCGCUC- ......((((((..-.(..(.(((.((((.((((((((((....)))))))).)))))).))).)..)(((........))).......))))))..- ( -37.30, z-score = -3.39, R) >droSim1.chr2R 15655737 96 + 19596830 AAGGAUGCGCAAUC-AGGACUCUGGCUUGCAGAUGAAAGGGAAACUUUUCAUACUCAAGGCAGCGGGCGCCGCUAAGUAGGCUACAAUUUUGCGCUC- ......((((((..-.(..(.(((.((((.((((((((((....)))))))).)))))).))).)..)(((........))).......))))))..- ( -37.30, z-score = -3.39, R) >consensus AAGGAUGCGCAAUC_AGGACUCUGGCUUGCAGAUGAAAGGGAAACUUUUCAUACUCAAGGCAGCGGGCGCCGCUAAGUAGGCUACAAUUUUGCGCUC_ ......((((((....(..(.(((.((((.((((((((((....)))))))).)))))).))).)..)(((........))).......))))))... (-25.76 = -27.52 + 1.75)


| Location | 16,998,675 – 16,998,772 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 90.34 |
| Shannon entropy | 0.18407 |
| G+C content | 0.49987 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -20.14 |
| Energy contribution | -20.28 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.506321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16998675 97 - 21146708 -GAGCGCAAAAUUGUAGCCUACUUAGCGGCGCCCGCUGCCUUGAGUAUGAAAAGUUUCACUUUCAUCUGCCAGCCAGAGUCCUGGAUUGCGCAUCCUU -..((((((....((((..(((((((((((....)))))..))))))(((((........))))).))))...((((....)))).))))))...... ( -30.20, z-score = -1.62, R) >droAna3.scaffold_13266 12922715 95 + 19884421 UUGGCA-AAAAGUGUAGCCUACUUAACUGCGCACGCUGUC-UGAAUGGGAAAAGUUUCCCUUUCAUCUGACUACCAUAGUCCU-GAUUACGCAUCCUU ...((.-....((((((.........))))))..(..(((-((((.(((((....))))).))))...(((((...)))))..-)))..)))...... ( -21.70, z-score = -1.25, R) >droEre2.scaffold_4845 11142911 96 - 22589142 -GAGCGCAAAAUUGUAGCCUACUUAGCGGCGCCCGCUGCCUUGAGUAUGAAAAGUUUCCCUUUCAUCUGCAGGACGGAGUCCU-GAUUGCGCAUCCUU -..((((((....((((..(((((((((((....)))))..))))))(((((........))))).))))(((((...)))))-..))))))...... ( -31.20, z-score = -1.81, R) >droYak2.chr2R 8943104 96 + 21139217 -GAGCGCAAAAUUGUAGCCUACUUAGUGGCGUCCGCUGCCUUGAGUAGGAAAAGUUUCCCUUUCAUCUGCAAGCCAGAGUCCU-GAUUGCGCAUCCUU -..((((((..(((((((((((((((.((((.....))))))))))))).((((.....))))...))))))..(((....))-).))))))...... ( -32.20, z-score = -2.95, R) >droSec1.super_9 328099 96 - 3197100 -GAGCGCAAAAUUGUAGCCUACUUAGCGGCGCCCGCUGCCUUGAGUAUGAAAAGUUUCCCUUUCAUCUGCAAGCCAGAGUCCU-GAUUGCGCAUCCUU -..((((((..((((((..(((((((((((....)))))..))))))(((((........))))).))))))..(((....))-).))))))...... ( -29.80, z-score = -2.37, R) >droSim1.chr2R 15655737 96 - 19596830 -GAGCGCAAAAUUGUAGCCUACUUAGCGGCGCCCGCUGCCUUGAGUAUGAAAAGUUUCCCUUUCAUCUGCAAGCCAGAGUCCU-GAUUGCGCAUCCUU -..((((((..((((((..(((((((((((....)))))..))))))(((((........))))).))))))..(((....))-).))))))...... ( -29.80, z-score = -2.37, R) >consensus _GAGCGCAAAAUUGUAGCCUACUUAGCGGCGCCCGCUGCCUUGAGUAUGAAAAGUUUCCCUUUCAUCUGCAAGCCAGAGUCCU_GAUUGCGCAUCCUU ...((((((..........(((((((((((....)))))..)))))).((((........)))).((((.....))))........))))))...... (-20.14 = -20.28 + 0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:51 2011