
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,485,291 – 3,485,385 |
| Length | 94 |
| Max. P | 0.579775 |

| Location | 3,485,291 – 3,485,385 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 67.16 |
| Shannon entropy | 0.59058 |
| G+C content | 0.52205 |
| Mean single sequence MFE | -27.85 |
| Consensus MFE | -12.65 |
| Energy contribution | -13.29 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.579775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3485291 94 - 23011544 --ACUCAGCGACACACCCUAAUCGGGCCAUGUCUUUGGAUCUCGGUCUUGGACUCU--GUUUUCGAGUU--UUCGAGAAACUAUGUGUGCUGGCUGCAAU------------------ --..(((((.((((((((.....)))(((......))).((((((.((((((....--...))))))..--.)))))).....)))))))))).......------------------ ( -28.40, z-score = -1.55, R) >droSim1.chr2L 3438597 94 - 22036055 --ACUCAGCGACACACCCUAAUCGGGCCAUGUCUUUGGAUCUCGGUCUUGGACUCU--GUGUUCGAGUU--UUCGAGAAACUAUGUGUGCUGGCUGCGAU------------------ --..(((((.((((((((.....)))(((......))).((((((.(((((((...--..)))))))..--.)))))).....)))))))))).......------------------ ( -32.70, z-score = -2.27, R) >droSec1.super_129 47253 94 - 51580 --ACUCAGCGACACACCCUAAUCGGGCCAUGUCUUUGGAUCUCGGUCUUGGACUCU--GUGUUCGAGUU--UUCGAGAAACUAUGUGUGCUGGCUGCGAU------------------ --..(((((.((((((((.....)))(((......))).((((((.(((((((...--..)))))))..--.)))))).....)))))))))).......------------------ ( -32.70, z-score = -2.27, R) >droYak2.chr2L 3473174 112 - 22324452 --ACUCAGCGACACACCCUAAUCGGGCCAUGUCUUUGGAUCUCGGUCUUGGACUUU--GUUUUCGAGUU--UUCGAGACACUACGUGUGCGAGUAUGUGUGCUGCUGCUGGCUGCGAU --...(((((.(((((.....(((.((.((((....(..((((((.((((((....--...))))))..--.))))))..).)))))).)))....))))).)))))........... ( -33.60, z-score = -0.20, R) >droEre2.scaffold_4929 3520021 97 - 26641161 --ACUCAGCGACACACCCUAAUCGGGCCAUGUCUUUGGAUCUCAGUCUUGGACUCU--GUUUUCGAGUU--UUCGAGACACUAUGUGUGCUGGCGGCUCCGAU--------------- --..(((((.((((((((.....)))(((......)))......(((((((((((.--......)))))--..))))))....))))))))))((....))..--------------- ( -29.60, z-score = -1.05, R) >droAna3.scaffold_12916 5532125 90 - 16180835 AUACUCAACGACACACCCUAAUCGGGCCAUGUCUUCGGGUCCGAAUCUCAAAUUUUUAAAGUCUCAGUCCGUUUCGGAGCCAGAGUCAGA---------------------------- ..((((...((((..(((.....)))...))))...((.((((((...........................)))))).)).))))....---------------------------- ( -22.63, z-score = -1.56, R) >dp4.chr4_group3 6427545 97 + 11692001 --ACCCAGCGACACACCCUAAUCUGGCUCUGGCUCUGGCUCU--GCCUCGGACUCU--AACUCGGACUUGGACUCUGGGACUGCGACUGCGACUUCGAUUUGA--------------- --.(.(((((.((..(((......(((...(((....)))..--)))..(((.(((--((.......))))).))))))..)))).))).)............--------------- ( -21.10, z-score = 0.27, R) >droPer1.super_1 3527674 97 + 10282868 --ACCCAGCGACACACCCUAAUCUGGCUCUGGCUCUGCCUCU--GCCUCGGACUCU--AACUCGGACUUGGACUCUGGGACUGCGACUGCGACUUCGAUUUGA--------------- --.(.(((((.((..(((......(((...(((...)))...--)))..(((.(((--((.......))))).))))))..)))).))).)............--------------- ( -22.10, z-score = -0.51, R) >consensus __ACUCAGCGACACACCCUAAUCGGGCCAUGUCUUUGGAUCUCGGUCUUGGACUCU__GUGUUCGAGUU__UUCGAGAAACUAUGUGUGCUGGCUGCAAU__________________ ....((((.((((..(((.....)))...)))).)))).((((((.....(((((.........)))))...))))))........................................ (-12.65 = -13.29 + 0.64)

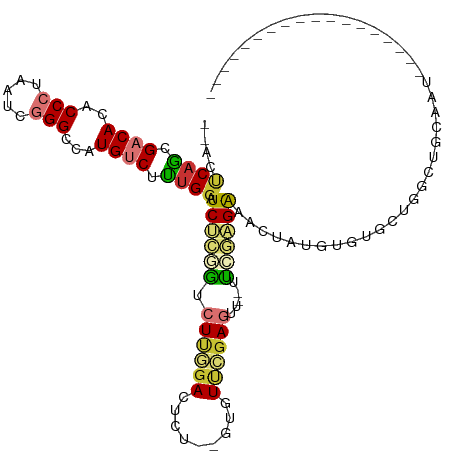
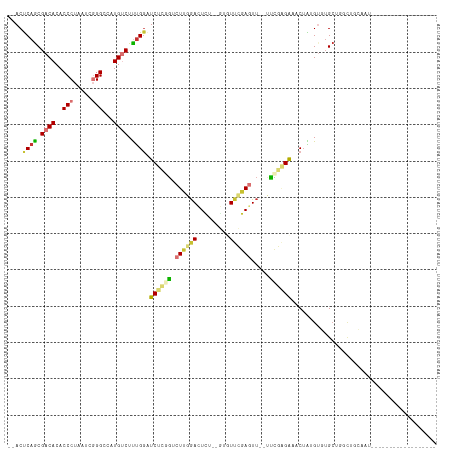
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:14:00 2011