
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,911,159 – 16,911,270 |
| Length | 111 |
| Max. P | 0.748247 |

| Location | 16,911,159 – 16,911,270 |
|---|---|
| Length | 111 |
| Sequences | 13 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.34 |
| Shannon entropy | 0.39005 |
| G+C content | 0.57173 |
| Mean single sequence MFE | -42.16 |
| Consensus MFE | -27.26 |
| Energy contribution | -27.22 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.748247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16911159 111 + 21146708 GGUGCAGGUACUCGGCAUGGAGUUAACAGCCAGUUGCGAGGGCGGCCACAAGUGGA---CUGGACGCCCUCCAACGCCUGUUUUCCGA---GCGCCAAGCGAACAUGUAAGUGCCAC- ......((((((..((((((.(..(((((...((((.(((((((.(((........---.))).)))))))))))..)))))..))..---((.....))...))))).))))))..- ( -44.20, z-score = -1.49, R) >droGri2.scaffold_15245 11359428 111 - 18325388 GGUGCAGGUUCUUGGCAUGGAGUUAACAACUAGUUGCGAGGGCGGUCAAAGAUGGA---CCGGAAGACCUCCAACGCCAGUUUAUCGC---ACAAAUAGUGGAUAUGUAAGUGUGCC- ((..((.....(((((....(((.....))).((((.((((.(((((.......))---))).....)))))))))))))..((((.(---((.....)))))))......))..))- ( -34.90, z-score = -1.35, R) >droMoj3.scaffold_6496 20688305 111 + 26866924 GCUGCAGGUUCUUGGCAUGGAGCUAACAGCUAGUUGUGAGGGCGGUCAGAGGUGGA---CCGGACGCCCUCCAACACCUGUUUAUCGC---GCAUCCAGUGAACAUGUAAGUGGCUC- ((..(..((((((((.(((..((.(((((...((((.(((((((.((...((....---)).)))))))))))))..)))))....))---.))))))).))))......)..))..- ( -43.20, z-score = -1.47, R) >droVir3.scaffold_12875 13392985 111 + 20611582 GGUGCAGGUUCUUGGCAUGGAGGUAACAGCCAGUUGCGAGGGCGGUCAAAGAUGGA---CUGGACGCCCUCCAACGCCUGUCUUCCGC---GCAAAUAGUGAACAUGUAAGUGCCUC- ((.((((((...((((.((.......))))))((((.(((((((..((........---.))..)))))))))))))))))...))((---((..((((....).)))..))))...- ( -42.40, z-score = -1.54, R) >droWil1.scaffold_181141 1509620 118 - 5303230 GAUUUAGGUUUUGGGCAUGGAGUUAACAGCCAGUUGUGAGGGCGGCCAAAAAUGGAAUGCUGGACGCCCUCCAACUCCUGUUUUCCGAACAGCACAAAGCGAACAUGUAAGUGUUGCC .......((((((.((.(((((..(((((..(((((.(((((((.(((............))).)))))))))))).))))))))))....)).)))))).(((((....)))))... ( -40.50, z-score = -1.84, R) >droPer1.super_31 681410 111 - 935084 GGCUCAGGUACUGGGCAUGGAGGUAACAGCCAGUUGCGAGGGCGGCCGCAAAUGGA---CCGGACGCCCUCCAACGCCUGUUUUCCGA---GCACCAAUCGAACAUGUAAGUGCCAC- ......((((((..(((((..((.(((((...((((.(((((((.(((........---.))).)))))))))))..)))))..))((---.......))...))))).))))))..- ( -46.20, z-score = -2.24, R) >dp4.chr3 17538867 111 + 19779522 GGCUCAGGUACUGGGCAUGGAGGUAACAGCCAGUUGCGAGGGCGGCCGCAAAUGGA---CCGGACGCCCUCCAACGCCUGUUUUCCGA---GCACCAAUCGAACAUGUAAGUGCCAC- ......((((((..(((((..((.(((((...((((.(((((((.(((........---.))).)))))))))))..)))))..))((---.......))...))))).))))))..- ( -46.20, z-score = -2.24, R) >droAna3.scaffold_13266 12824408 111 - 19884421 GGUUCAGGUACUGGGCAUGGAGUUAACAGCCAGUUGCGAGGGUGGCCAAAAGUGGA---CUGGACGCCCUCCAACGCCUGUUUUCCGA---GCACCAAACGAACAUGUAAGUGCCAC- ......((((((((((.(((((..(((((...((((.(((((((.(((........---.))).)))))))))))..)))))))))).---)).))..((......)).))))))..- ( -42.90, z-score = -1.99, R) >droYak2.chr2R 8853634 111 - 21139217 GGUGCAGGUACUCGGCAUGGAGUUAACAGCCAGUUGCGAGGGCGGCCACAAGUGGA---CUGGACGCCCUCCAACGCCUGUUUUCCGA---GCGCCAAGCGAACAUGUAAGCAGCAC- .((((.((..(((((.........(((((...((((.(((((((.(((........---.))).)))))))))))..)))))..))))---)..))..((..........)).))))- ( -44.20, z-score = -1.61, R) >droEre2.scaffold_4845 11055167 111 + 22589142 GGUGCAGGUACUCGGCAUGGAGUUAACAGCCAGUUGCGAGGGCGGCCACAAGUGGA---CUGGACGCCCUCCAACGCCUGUUUUCCGA---GCGCCAAGCGAACAUGUAAGUGCCAC- ......((((((..((((((.(..(((((...((((.(((((((.(((........---.))).)))))))))))..)))))..))..---((.....))...))))).))))))..- ( -44.20, z-score = -1.49, R) >droSec1.super_9 241822 111 + 3197100 GGUGCAGGUACUCGGCAUGGAGUUAACAGCCAGUUGCGAGGGCGGCCACAAGUGGA---CUGGACGCCCUCCAACGCCUGUUUUCCGA---GCGCCAAGCGAACAUGUAAGUGCCAC- ......((((((..((((((.(..(((((...((((.(((((((.(((........---.))).)))))))))))..)))))..))..---((.....))...))))).))))))..- ( -44.20, z-score = -1.49, R) >droSim1.chr2R 15566439 111 + 19596830 GGUGCAGGUACUCGGCAUGGAGUUAACAGCCAGUUGCGAGGGCGGCCACAAGUGGA---CUGGACGCCCUCCAACGCCUGUUUUCCGA---GCGCCAAGCGAACAUGUAAGUGCCAC- ......((((((..((((((.(..(((((...((((.(((((((.(((........---.))).)))))))))))..)))))..))..---((.....))...))))).))))))..- ( -44.20, z-score = -1.49, R) >triCas2.ChLG7 8668502 110 + 17478683 GGUUAAGGUUUUGGGAAUGGAAAUGACAGCCAGUUGUGAAGGCGGGAAGAAAUGGU------CAAGGCCUGGAUCACCAGUUCUACCAAC-ACCCUCACCGACUCCUUCAGAACCAG- ......((((((((((.(((........(((.........)))(((......((((------..((..((((....))))..))))))..-.)))...)))..)))..)))))))..- ( -30.80, z-score = 0.33, R) >consensus GGUGCAGGUACUCGGCAUGGAGUUAACAGCCAGUUGCGAGGGCGGCCACAAGUGGA___CUGGACGCCCUCCAACGCCUGUUUUCCGA___GCACCAAGCGAACAUGUAAGUGCCAC_ (((.(((((....(((.((.......))))).((((.(((((((.(((............))).)))))))))))))))).....((............))...........)))... (-27.26 = -27.22 + -0.04)

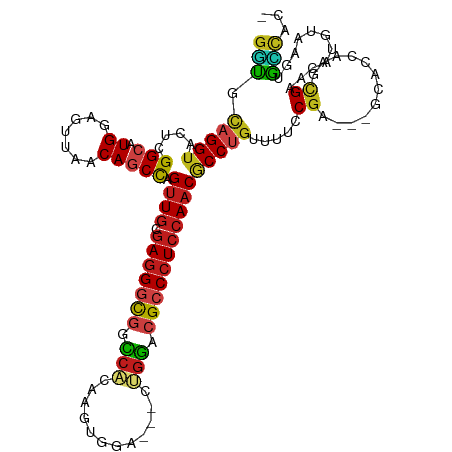
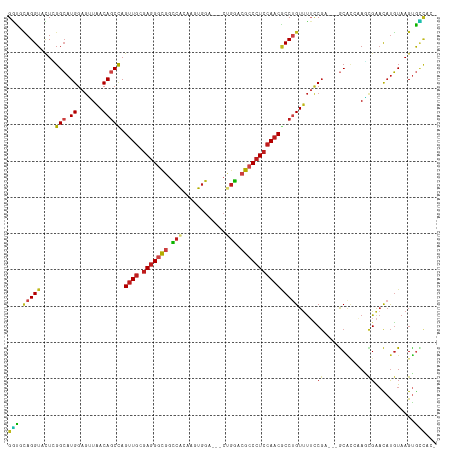
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:45 2011