
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,869,555 – 16,869,674 |
| Length | 119 |
| Max. P | 0.650488 |

| Location | 16,869,555 – 16,869,674 |
|---|---|
| Length | 119 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.85 |
| Shannon entropy | 0.75071 |
| G+C content | 0.49264 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -11.37 |
| Energy contribution | -11.03 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.61 |
| Mean z-score | -0.53 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.650488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
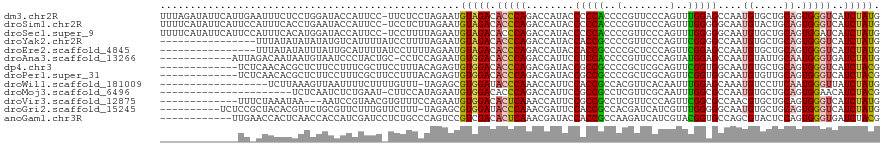
>dm3.chr2R 16869555 119 + 21146708 UUUAGAUAUUCAUUGAAUUUCUCCUGGAUACCAUUCC-UUCUCCUAGAAUGUAGACACCCAGACCAUACCCCCACCCCGUUCCCAGUUUGGAGCCAAUGUGCUGCAGUGGGUCAUCUAUG ..(((((((((((((........((((.(((.((((.-........))))))).....))))..........(((...(((((......)))))....)))...)))))))).))))).. ( -23.60, z-score = -0.17, R) >droSim1.chr2R 15524194 119 + 19596830 UUUUCAUAUUCAUUCCAUUUCACCUGAAUACCAUUCC-UCCUCUUAGAAUGUAGACACCCAGACCAUACCCCCACCCCGUUCCCAGUUUGGGGGCAAUGUACUGCAGUGGGUCAUCUAUG ......((((((............)))))).(((((.-........)))))((((.(((((........(((((..(........)..)))))(((......)))..)))))..)))).. ( -23.60, z-score = -0.07, R) >droSec1.super_9 200271 119 + 3197100 UUUUCAUAUUCAUUCCAUUUCACAUGGAUACCAUUCC-UCCUUUUAGAAUGUAGACACCCAGACCAUACCCCCACCCCGUUCCCAGUUUGGGGGCAAUGUGCUGCAGUGGAUCAUCUAUG ......................(((((((..(((((.-........)))))....(((.(((..(((.((((((..(........)..))))))..)))..)))..)))....))))))) ( -26.80, z-score = -0.88, R) >droYak2.chr2R 8811702 104 - 21139217 ----------------UUUAUAUAUAUAUGUCAUUUUAUCCUUUUAGAAUGUAGACACCCAGACCAUACCACCGCCCCGUUCCCAGUUCGGGGCCAAUGUGCUGCAGUGGGUCAUCUAUG ----------------...............(((((((......)))))))((((.(((((...(((((....((((((.........))))))....))).))...)))))..)))).. ( -25.70, z-score = -1.36, R) >droEre2.scaffold_4845 11013092 104 + 22589142 ----------------UUUAUAUAUUUAUUGCAUUUUAUCCUUUUAGAAUGUAGACACCCAGACCAUACCACCGCCCCGCUCCCAGUUCGGAGCCAAUGUGCUGCAGUGGGUCAUCUAUG ----------------............((((((((((......)))))))))).......((((.(((((.(((...(((((......)))))....))).))..)))))))....... ( -25.10, z-score = -2.38, R) >droAna3.scaffold_13266 16172780 107 - 19884421 ------------AUUAGACAAUAAUGUAAUCCCUACUGC-CCUCCAGAAUGUGGACACCCAGACCAUUCCUCCACCCCGUUCCCAGUAUGGAGCCAAUGUAUUGCAAUGGGUGAUCUAUG ------------..((((...........((((..(((.-....)))...).)))((((((...((((.(((((..(........)..)))))..))))........)))))).)))).. ( -20.40, z-score = 0.34, R) >dp4.chr3 17488970 107 + 19779522 -------------UCUCAACACGCUCUUCCUUUCGCUUCCUUUACAGAGUGUGGACACCCAGACGAUACCGCCGCCCCGCUCGCAGUUCGGUGGCAAUGUGCUGCAGUGGGUCAUCUACG -------------......((((((((..................))))))))(((.((((((((...((((((..(.(....).)..))))))...))).)))....)))))....... ( -28.07, z-score = 0.48, R) >droPer1.super_31 632472 107 - 935084 -------------UCUCAACACGCUCUUCCUUUCGCUUCCUUUACAGAGUGUGGACACCCAGACGAUACCGCCGCCCCGCUCGCAGUUCGGUGGCAAUGUGUUGCAGUGGGUCAUCUACG -------------......((((((((..................))))))))...(((((..((((((.((((((..((.....))..))))))...))))))...)))))........ ( -32.27, z-score = -0.98, R) >droWil1.scaffold_181009 2212135 101 + 3585778 ------------------UCUUAAAGUUAAUUUUCUUUUGUUU-UAGAGCGUGGAUACCCAAACCAUUCCACCGCCACGUUCACAAUUUGGAGCAAAUGUCCUUCAAUGGGUUAUCUAUG ------------------......((.((((.....(((((((-((((((((((....................))))))))......)))))))))...((......)))))).))... ( -16.65, z-score = -0.03, R) >droMoj3.scaffold_6496 20633884 97 + 26866924 ----------------------UCUCAAUCUCUGAAU-CUUCCAUAGAAUGUGGACACCCAGACCAUUCCGCCGCCUCGUUCGCAAUUUGGCGCCAAUGUGCUGCAGUGGAACAUCUACG ----------------------........((((...-..(((((.....)))))....))))...((((((.((..(((((((......)))..))))....)).))))))........ ( -20.90, z-score = -0.03, R) >droVir3.scaffold_12875 13336632 104 + 20611582 -------------UUUCUAAAUAA---AAUCCGUAACGUGUUUCCAGAAUGUGGACACUCAAACCAUUCCGCCGCCUCGUUCCCAGUUCGGCGCCAACGUGCUGCAGUGGGUCAUCUAUG -------------...........---....((((..(((((..(.....)..)))))....................(..((((.(.((((((....)))))).).)))).)...)))) ( -21.50, z-score = -0.04, R) >droGri2.scaffold_15245 11310344 109 - 18325388 ----------UCUCCGCUACACGUUCUGCGUUCUUUGUUCUUU-UAGAGCGUGGAUACCCAAACGAUUCCACCGCCACGAUCAUCGUUUGGGGGCAAUGUGCUGCAGUGGGUCAUCUAUG ----------..((((((.(((((((..((((((..(....).-.))))))..))..((((((((((...............))))))))))....)))))....))))))......... ( -34.46, z-score = -1.77, R) >anoGam1.chr3R 15219934 108 + 53272125 ------------UUGAACCACUCAACCACCAUCGAUCCUCUGCCCAGUCCGUCGACACUCAAACGAUACCACCGCCAAGAUCAUCGUACGGUGCCAGCGUACUCCAGUGGGUGAUCUACG ------------((((.....)))).((((.(((((...((....))...)))))((((...(((....(((((....(.....)...)))))....))).....))))))))....... ( -22.80, z-score = -0.00, R) >consensus _____________UC_AUUAAUCAUCUAUAUCAUUCU_UCUUUCUAGAAUGUGGACACCCAGACCAUACCACCGCCCCGUUCCCAGUUUGGAGCCAAUGUGCUGCAGUGGGUCAUCUAUG ..................................................(((((.(((((........(((((..(........)..)))))....((.....)).)))))..))))). (-11.37 = -11.03 + -0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:43 2011